Difference between revisions of "Main Page/Featured article of the week/2017"
Shawndouglas (talk | contribs) (Added last week's article of the week.) |
Shawndouglas (talk | contribs) m (Text replacement - "\[\[Satya Sistemas Ltda.(.*)" to "[[Vendor:Satya Sistemas Ltda.$1") |
||
| (46 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{ombox | {{ombox | ||
| type = notice | | type = notice | ||
| text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - 2017 | | text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - 2017 - [[Main Page/Featured article of the week/2018|2018]] - [[Main Page/Featured article of the week/2019|2019]] - [[Main Page/Featured article of the week/2020|2020]] - [[Main Page/Featured article of the week/2021|2021]] - [[Main Page/Featured article of the week/2022|2022]] - [[Main Page/Featured article of the week/2023|2023]] - [[Main Page/Featured article of the week/2024|2024]] | ||
}} | }} | ||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 3–9:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 25–31:</h2> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Tremouilhac JOfChemoinfo2017 9.gif|240px]]</div> | |||
'''"[[Journal:Chemotion ELN: An open-source electronic lab notebook for chemists in academia|Chemotion ELN: An open-source electronic lab notebook for chemists in academia]]"''' | |||
The development of an [[electronic laboratory notebook]] (ELN) for researchers working in the field of chemical sciences is presented. The web-based application is available as open-source software that offers modern solutions for chemical researchers. The [[Chemotion ELN]] is equipped with the basic functionalities necessary for the acquisition and processing of chemical data, in particular work with molecular structures and calculations based on molecular properties. The ELN supports planning, description, storage, and management for the routine work of organic chemists. It also provides tools for communicating and sharing the recorded research data among colleagues. Meeting the requirements of a state-of-the-art research infrastructure, the ELN allows the search for molecules and reactions not only within the user’s data but also in conventional external sources as provided by SciFinder and PubChem. The presented development makes allowance for the growing dependency of scientific activity on the availability of digital [[information]] by providing open- source instruments to record and reuse research data. The current version of the ELN has been used for over half of a year in our chemistry research group, serving as a common infrastructure for chemistry research and enabling chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms. ('''[[Journal:Chemotion ELN: An open-source electronic lab notebook for chemists in academia|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 18–24:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Mayernik BigDataSoc2017 4-2.gif|240px]]</div> | |||
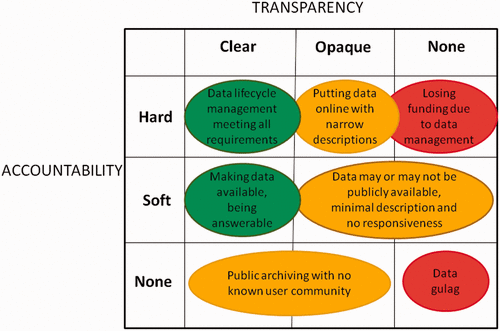
'''"[[Journal:Open data: Accountability and transparency|Open data: Accountability and transparency]]"''' | |||
The movements by national governments, funding agencies, universities, and research communities toward “open data” face many difficult challenges. In high-level visions of open data, researchers’ data and metadata practices are expected to be robust and structured. The integration of the internet into scientific institutions amplifies these expectations. When examined critically, however, the data and metadata practices of scholarly researchers often appear incomplete or deficient. The concepts of “accountability” and “transparency” provide insight in understanding these perceived gaps. Researchers’ primary accountabilities are related to meeting the expectations of research competency, not to external standards of data deposition or metadata creation. Likewise, making data open in a transparent way can involve a significant investment of time and resources with no obvious benefits. This paper uses differing notions of accountability and transparency to conceptualize “open data” as the result of ongoing achievements, not one-time acts. ('''[[Journal:Open data: Accountability and transparency|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 11–17:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Husen DataSciJourn2017 16-1.png|240px]]</div> | |||
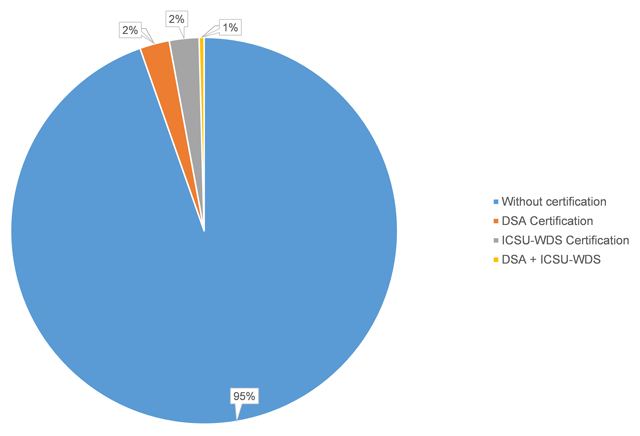
'''"[[Journal:Recommended versus certified repositories: Mind the gap|Recommended versus certified repositories: Mind the gap]]"''' | |||
Researchers are increasingly required to make research data publicly available in data repositories. Although several organizations propose criteria to recommend and evaluate the quality of data repositories, there is no consensus of what constitutes a good data repository. In this paper, we investigate, first, which data repositories are recommended by various stakeholders (publishers, funders, and community organizations) and second, which repositories are certified by a number of organizations. We then compare these two lists of repositories, and the criteria for recommendation and certification. We find that criteria used by organizations recommending and certifying repositories are similar, although the certification criteria are generally more detailed. We distill the lists of criteria into seven main categories: “Mission,” “Community/Recognition,” “Legal and Contractual Compliance,” “Access/Accessibility,” “Technical Structure/Interface,” “Retrievability,” and “Preservation.” Although the criteria are similar, the lists of repositories that are recommended by the various agencies are very different. Out of all of the recommended repositories, less than six percent obtained certification. As certification is becoming more important, steps should be taken to decrease this gap between recommended and certified repositories, and ensure that certification standards become applicable, and applied, to the repositories which researchers are currently using. ('''[[Journal:Recommended versus certified repositories: Mind the gap|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 4–10:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab2 Matthews JPathInfo2017 8.jpg|240px]]</div> | |||
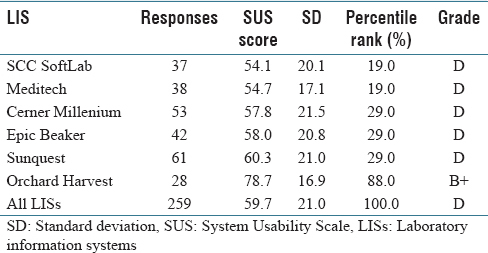
'''"[[Journal:Usability evaluation of laboratory information systems|Usability evaluation of laboratory information systems]]"''' | |||
Numerous studies have revealed widespread clinician frustration with the usability of [[electronic health record]]s (EHRs) that is counterproductive to adoption of EHR systems to meet the aims of healthcare reform. With poor system usability comes increased risk of negative unintended consequences. Usability issues could lead to user error and workarounds that have the potential to compromise patient safety and negatively impact the quality of care. While there is ample research on EHR usability, there is little [[information]] on the usability of [[laboratory information system]]s (LIS). Yet, an LIS facilitates the timely provision of a great deal of the information needed by physicians to make patient care decisions. Medical and technical advances in genomics that require processing of an increased volume of complex [[laboratory]] data further underscore the importance of developing a user-friendly LIS. This study aims to add to the body of knowledge on LIS usability. ('''[[Journal:Usability evaluation of laboratory information systems|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 27–December 3:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Martin LIBER2017 27-1.jpg|240px]]</div> | |||
'''"[[Journal:Data management: New tools, new organization, and new skills in a French research institute|Data management: New tools, new organization, and new skills in a French research institute]]"''' | |||
In the context of e-science and open access, visibility and impact of scientific results and data have become important aspects for spreading [[information]] to users and to the society in general. The objective of this general trend of the economy is to feed the innovation process and create economic value. In our institute, the French National Research Institute of Science and Technology for Environment and Agriculture, Irstea, the department in charge of scientific and technical information, with the help of other professionals (scientists, IT professionals, ethics advisors, etc.), has recently developed suitable services for researchers and their data management needs in order to answer European recommendations for open data. This situation has demanded a review of the different workflows between databases, questioning the organizational aspects among skills, occupations, and departments in the institute. In fact, data management involves all professionals and researchers assessing their workflows together. ('''[[Journal:Data management: New tools, new organization, and new skills in a French research institute|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 20–26:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Bellgard FrontPubHealth2017 5.jpg|240px]]</div> | |||
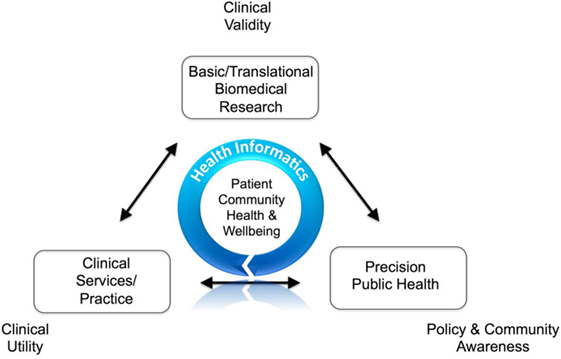
'''"[[Journal:Comprehending the health informatics spectrum: Grappling with system entropy and advancing quality clinical research|Comprehending the health informatics spectrum: Grappling with system entropy and advancing quality clinical research]]"''' | |||
Clinical research is complex. The knowledge base is [[information]]- and data-rich, where value and success depend upon focused, well-designed connectivity of systems achieved through stakeholder collaboration. Quality data, information, and knowledge must be utilized in an effective, efficient, and timely manner to affect important clinical decisions and communicate health prevention strategies. In recent decades, it has become apparent that information communication technology (ICT) solutions potentially offer multidimensional opportunities for transforming health care and clinical research. However, it is also recognized that successful utilization of ICT in improving patient care and health outcomes depends on a number of factors such as the effective integration of diverse sources of health data; how and by whom quality data are captured; reproducible methods on how data are interrogated and reanalyzed; robust policies and procedures for data privacy, security and access; usable consumer and clinical user interfaces; effective diverse stakeholder engagement; and navigating the numerous eclectic and non-interoperable legacy proprietary health ICT solutions in [[hospital]] and clinic environments. This is broadly termed [[health informatics]] (HI). ('''[[Journal:Comprehending the health informatics spectrum: Grappling with system entropy and advancing quality clinical research|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 13–19:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Huang ICTExpress2017 3-2.jpg|240px]]</div> | |||
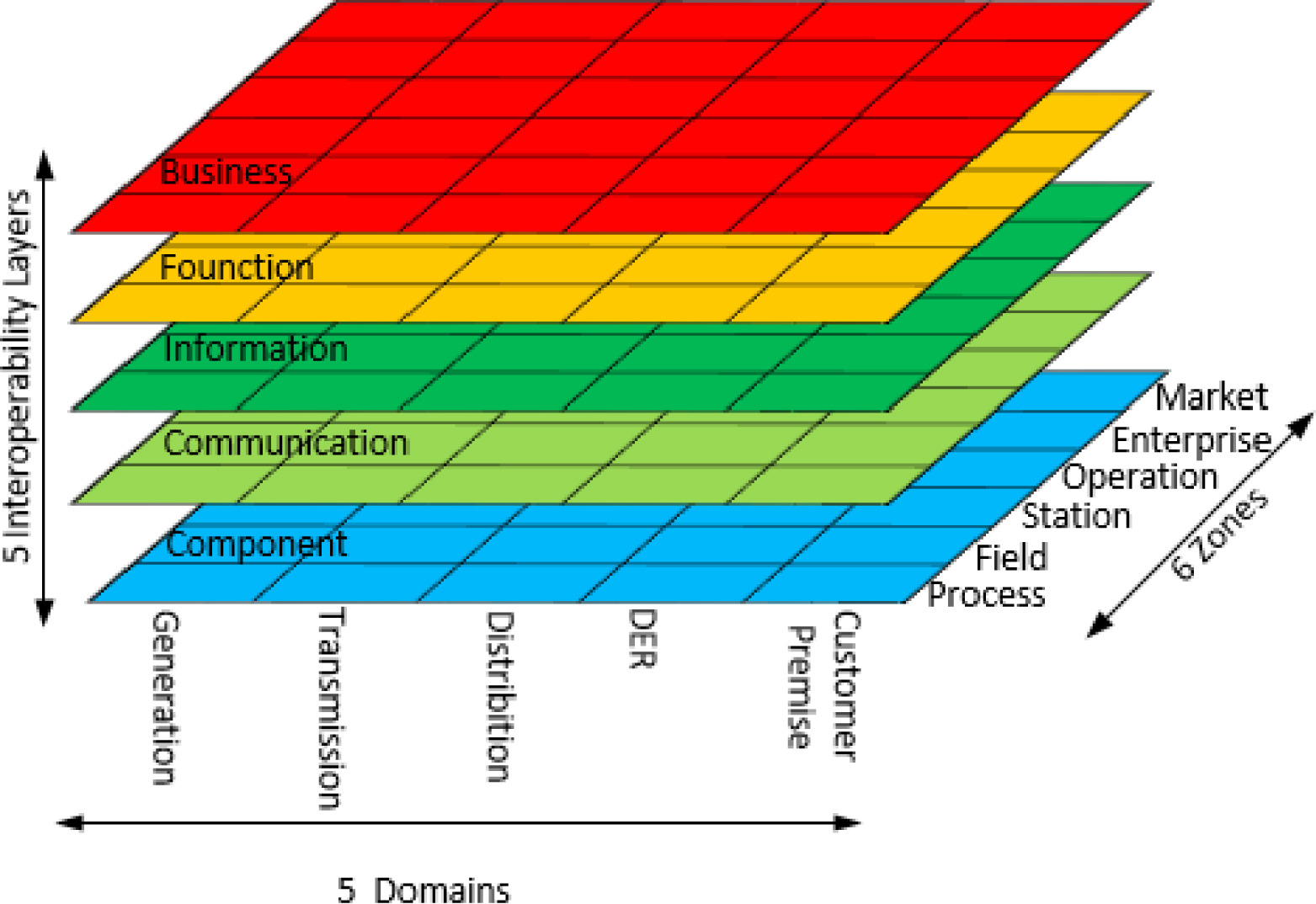
'''"[[Journal:Energy informatics: Fundamentals and standardization|Energy informatics: Fundamentals and standardization]]"''' | |||
Based on international standardization and power utility practices, this paper presents a preliminary and systematic study on the field of energy [[informatics]] and analyzes boundary expansion of information and energy systems, and the convergence of energy systems and ICT. A comprehensive introduction of the fundamentals and standardization of energy informatics is provided, and several key open issues are identified. | |||
With the changing of global climate and a world energy shortage, a smooth transition from conventional fossil fuel-based energy supplies to renewable energy sources is critical for the sustainable development of human society. Meanwhile, the energy domain is experiencing a paradigmatic change by integrating conventional energy systems with advanced [[information]] and communication technologies (ICT), which poses new challenges to the efficient operation and design of energy systems. ('''[[Journal:Energy informatics: Fundamentals and standardization|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 6–12:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Vaas PeerJCompSci2016 2.jpg|240px]]</div> | |||
'''"[[Journal:Electronic laboratory notebooks in a public–private partnership|Electronic laboratory notebooks in a public–private partnership]]"''' | |||
This report shares the experience during selection, implementation and maintenance phases of an [[electronic laboratory notebook]] (ELN) in a public–private partnership project and comments on users' feedback. In particular, we address which time constraints for roll-out of an ELN exist in granted projects and which benefits and/or restrictions come with out-of-the-box solutions. We discuss several options for the implementation of support functions and potential advantages of open-access solutions. Connected to that, we identified willingness and a vivid culture of data sharing as the major item leading to success or failure of collaborative research activities. The feedback from users turned out to be the only angle for driving technical improvements, but also exhibited high efficiency. Based on these experiences, we describe best practices for future projects on implementation and support of an ELN supporting a diverse, multidisciplinary user group based in academia, NGOs, and/or for-profit corporations located in multiple time zones. ('''[[Journal:Electronic laboratory notebooks in a public–private partnership|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 30–November 5:</h2> | |||
'''"[[Journal:Laboratory information system – Where are we today?|Laboratory information system – Where are we today?]]"''' | |||
Wider implementation of [[laboratory information system]]s (LIS) in [[Clinical laboratory|clinical laboratories]] in Serbia was initiated 10 years ago. The first LIS in the Railway Health Care Institute was implemented nine years ago. Before the LIS was initiated, manual admission procedures limited daily output of patients. Moreover, manual entering of patient data and ordering tests on analyzers was problematic and time-consuming. After completing tests, [[laboratory]] personnel had to write results in a patient register (with potential errors) and provide invoices for health insurance organizations. The first LIS brought forward some advantages with regards to these obstacles, but it also showed various weaknesses. These can be summarized as rigidity of the system and inability to fulfill user expectation. After four years of use, we replaced this system with another LIS. Hence, the main aim of this paper is to evaluate the advantages of using LIS in the Railway Health Care Institute's laboratory and also to discuss further possibilities for its application. ('''[[Journal:Laboratory information system – Where are we today?|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 23–29:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Deliberato JMIRMedInfo2017 5-3.png|240px]]</div> | |||
'''"[[Journal:Clinical note creation, binning, and artificial intelligence|Clinical note creation, binning, and artificial intelligence]]"''' | |||
The creation of medical notes in software applications poses an intrinsic problem in workflow as the technology inherently intervenes in the processes of collecting and assembling [[information]], as well as the production of a data-driven note that meets both individual and healthcare system requirements. In addition, the note writing applications in currently available [[electronic health record]]s (EHRs) do not function to support decision making to any substantial degree. We suggest that artificial intelligence (AI) could be utilized to facilitate the workflows of the data collection and assembly processes, as well as to support the development of personalized, yet data-driven assessments and plans. ('''[[Journal:Clinical note creation, binning, and artificial intelligence|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 16–22:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Panahiazar JofBiomedInformatics2017 72-8.jpg|240px]]</div> | |||
'''"[[Journal:Predicting biomedical metadata in CEDAR: A study of Gene Expression Omnibus (GEO)|Predicting biomedical metadata in CEDAR: A study of Gene Expression Omnibus (GEO)]]"''' | |||
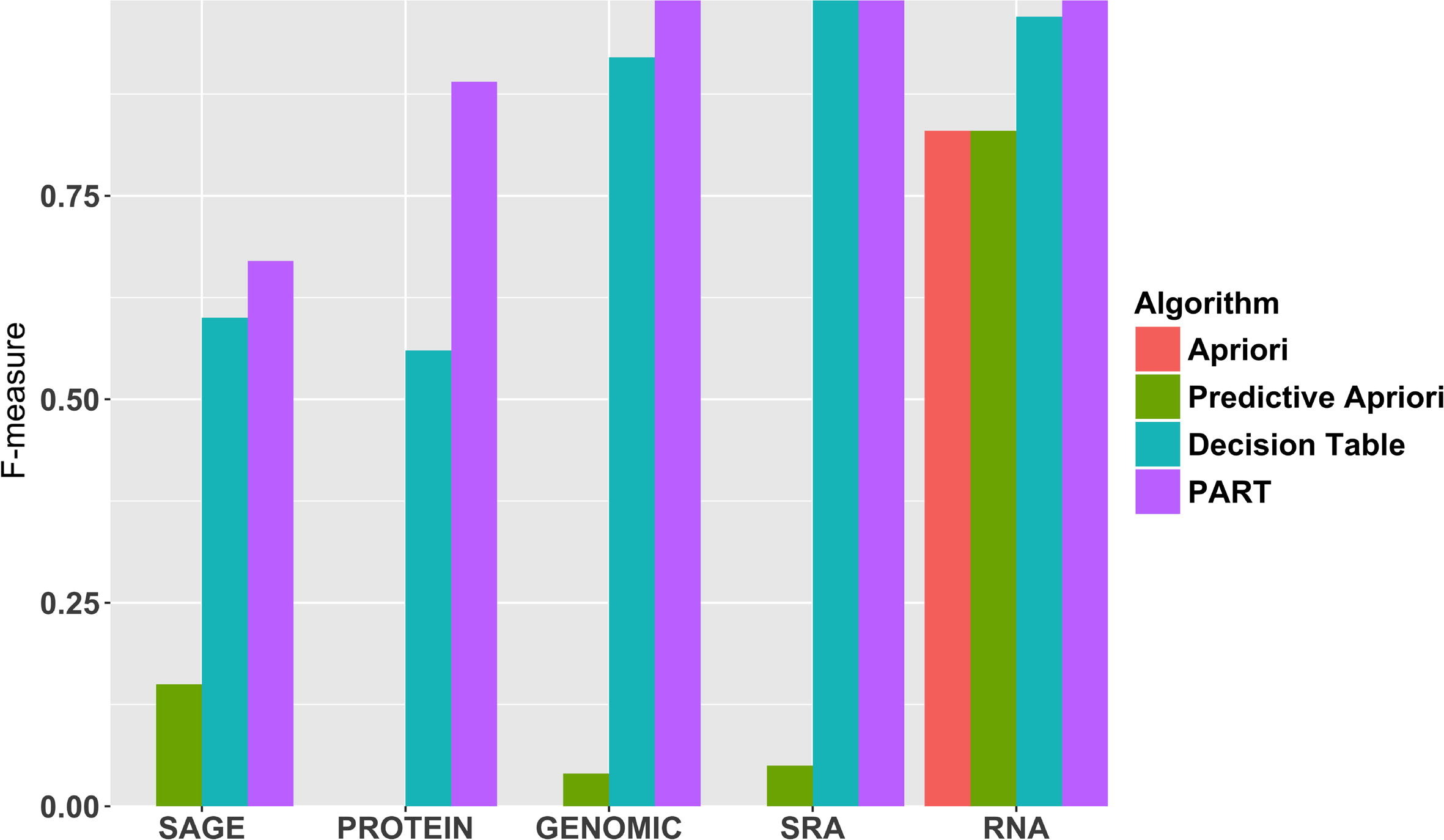
A crucial and limiting factor in data reuse is the lack of accurate, structured, and complete descriptions of data, known as metadata. Towards improving the quantity and quality of metadata, we propose a novel metadata prediction framework to learn associations from existing metadata that can be used to predict metadata values. We evaluate our framework in the context of experimental metadata from the Gene Expression Omnibus (GEO). We applied four rule mining algorithms to the most common structured metadata elements (sample type, molecular type, platform, label type and organism) from over 1.3 million GEO records. We examined the quality of well supported rules from each algorithm and visualized the dependencies among metadata elements. Finally, we evaluated the performance of the algorithms in terms of accuracy, precision, recall, and F-measure. We found that PART is the best algorithm outperforming Apriori, Predictive Apriori, and Decision Table. | |||
All algorithms perform significantly better in predicting class values than the majority vote classifier. We found that the performance of the algorithms is related to the dimensionality of the GEO elements. ('''[[Journal:Predicting biomedical metadata in CEDAR: A study of Gene Expression Omnibus (GEO)|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 9–15:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Tsur BioDataMining2017 10.gif|240px]]</div> | |||
'''"[[Journal:Rapid development of entity-based data models for bioinformatics with persistence object-oriented design and structured interfaces|Rapid development of entity-based data models for bioinformatics with persistence object-oriented design and structured interfaces]]"''' | |||
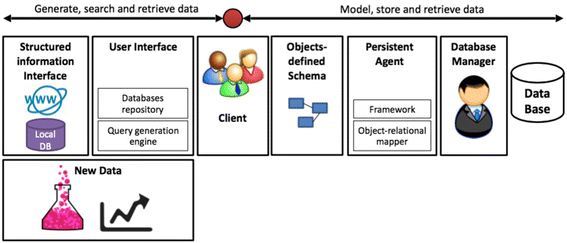
Databases are imperative for research in [[bioinformatics]] and computational biology. Current challenges in database design include data heterogeneity and context-dependent interconnections between data entities. These challenges drove the development of unified data interfaces and specialized databases. The curation of specialized databases is an ever-growing challenge due to the introduction of new data sources and the emergence of new relational connections between established datasets. Here, an open-source framework for the curation of specialized databases is proposed. The framework supports user-designed models of data encapsulation, object persistence and structured interfaces to local and external data sources such as MalaCards, Biomodels and the National Center for Biotechnology Information (NCBI) databases. The proposed framework was implemented using Java as the development environment, EclipseLink as the data persistence agent and Apache Derby as the database manager. Syntactic analysis was based on J3D, jsoup, Apache Commons and w3c.dom open libraries. Finally, a construction of a specialized database for aneurysm-associated vascular diseases is demonstrated. This database contains three-dimensional geometries of aneurysms, patients' clinical information, articles, biological models, related diseases and our recently published model of aneurysms’ risk of rapture. The framework is available at: http://nbel-lab.com. ('''[[Journal:Rapid development of entity-based data models for bioinformatics with persistence object-oriented design and structured interfaces|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 2–8:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Clay CancerInformatics2017 16.png|240px]]</div> | |||
'''"[[Journal:Bioinformatics education in pathology training: Current scope and future direction|Bioinformatics education in pathology training: Current scope and future direction]]"''' | |||
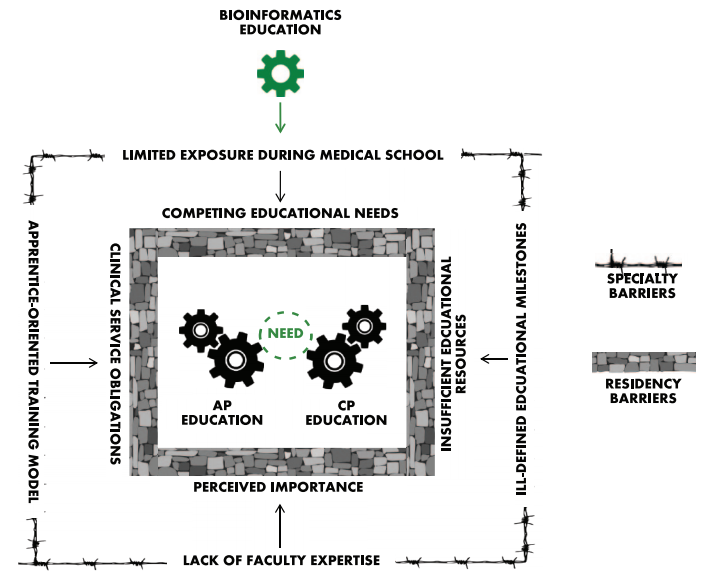
Training [[Anatomical pathology|anatomic]] and [[clinical pathology]] residents in the principles of [[bioinformatics]] is a challenging endeavor. Most residents receive little to no formal exposure to bioinformatics during medical education, and most of the pathology training is spent interpreting [[histopathology]] slides using light microscopy or focused on laboratory regulation, management, and interpretation of discrete [[laboratory]] data. At a minimum, residents should be familiar with data structure, data pipelines, data manipulation, and data regulations within [[Clinical laboratory|clinical laboratories]]. Fellowship-level training should incorporate advanced principles unique to each subspecialty. Barriers to bioinformatics education include the clinical apprenticeship training model, ill-defined educational milestones, inadequate faculty expertise, and limited exposure during medical training. Online educational resources, case-based learning, and incorporation into molecular genomics education could serve as effective educational strategies. Overall, pathology bioinformatics training can be incorporated into pathology resident curricula, provided there is motivation to incorporate institutional support, educational resources, and adequate faculty expertise. ('''[[Journal:Bioinformatics education in pathology training: Current scope and future direction|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 25–October 1:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Faria-Campos BMCBioinformatics2015 16-S19.jpg|240px]]</div> | |||
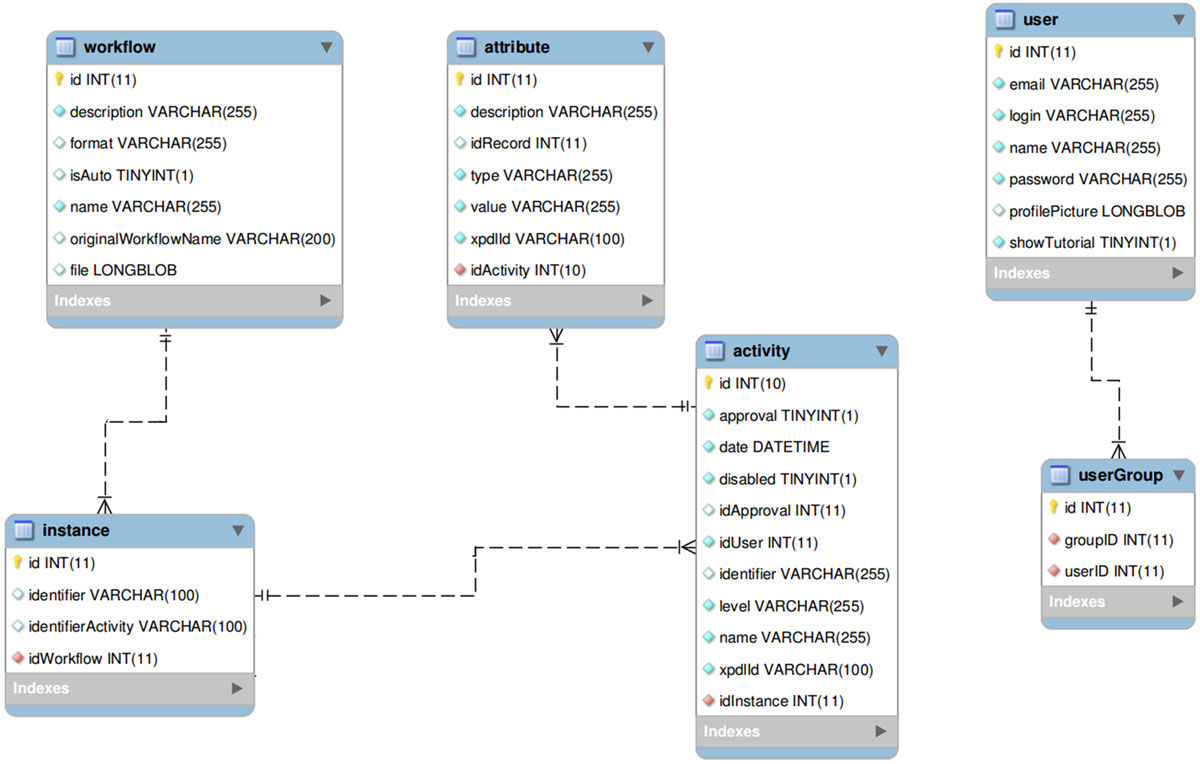
'''"[[Journal:FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data|FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data]]"''' | |||
Cytotoxicity assays have been used by researchers to screen for cytotoxicity in compound libraries. Researchers can either look for cytotoxic compounds or screen "hits" from initial high-throughput drug screens for unwanted cytotoxic effects before investing in their development as a pharmaceutical. These assays may be used as an alternative to animal experimentation and are becoming increasingly important in modern laboratories. However, the execution of these assays in large-scale and different laboratories requires, among other things, the management of protocols, reagents, and cell lines used, as well as the data produced, which can be a challenge. The management of all this information is greatly improved by the utilization of computational tools to save time and guarantee quality. However, a tool that performs this task designed specifically for cytotoxicity assays is not yet available. | |||
In this work, we have used a workflow based LIMS — [[Vendor:Satya Sistemas Ltda.#Flux2|the Flux system]] — and the Together Workflow Editor as a framework to develop FluxCTTX, a tool for management of data from cytotoxicity assays performed at different laboratories. ('''[[Journal:FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 18–24:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Shin JofPathInformatics2017 8.jpg|240px]]</div> | |||
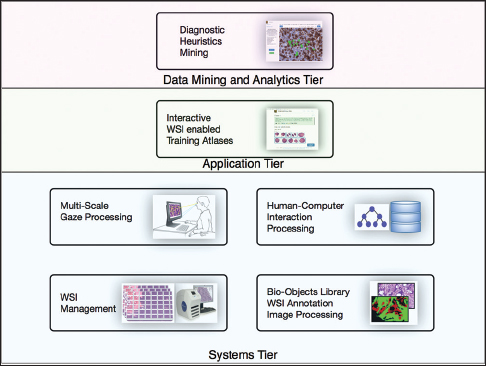
'''"[[Journal:PathEdEx – Uncovering high-explanatory visual diagnostics heuristics using digital pathology and multiscale gaze data|PathEdEx – Uncovering high-explanatory visual diagnostics heuristics using digital pathology and multiscale gaze data]]"''' | |||
Visual heuristics of pathology diagnosis is a largely unexplored area where reported studies only provided a qualitative insight into the subject. Uncovering and quantifying pathology visual and non-visual diagnostic patterns have great potential to improve clinical outcomes and avoid diagnostic pitfalls. | |||
Here, we present PathEdEx, an [[informatics]] computational framework that incorporates whole-slide digital pathology imaging with multiscale gaze-tracking technology to create web-based interactive pathology educational atlases and to datamine visual and non-visual diagnostic heuristics. | |||
We demonstrate the capabilities of PathEdEx for mining visual and non-visual diagnostic heuristics using the first PathEdEx volume of a [[Clinical pathology#Sub-specialties|hematopathology]] atlas. We conducted a quantitative study on the time dynamics of zooming and panning operations utilized by experts and novices to come to the correct diagnosis. ('''[[Journal:PathEdEx – Uncovering high-explanatory visual diagnostics heuristics using digital pathology and multiscale gaze data|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 11–17:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Kanza JofCheminformatics2017 9.gif|240px]]</div> | |||
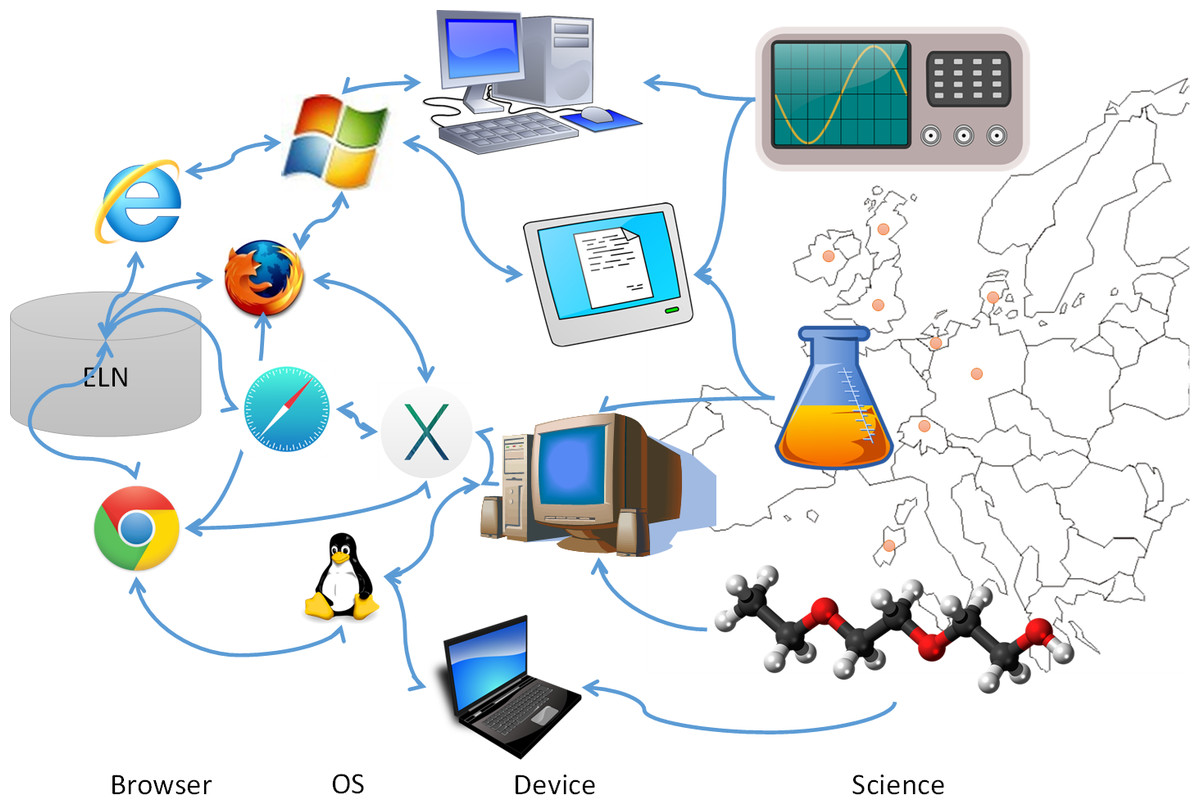
'''"[[Journal:Electronic lab notebooks: Can they replace paper|Electronic lab notebooks: Can they replace paper?]]"''' | |||
Despite the increasingly digital nature of society, there are some areas of research that remain firmly rooted in the past; in this case the [[laboratory notebook]], the last remaining paper component of an experiment. Countless [[electronic laboratory notebook]]s (ELNs) have been created in an attempt to digitize record keeping processes in the lab, but none of them have become a "key player" in the ELN market, due to the many adoption barriers that have been identified in previous research and further explored in the user studies presented here. The main issues identified are the cost of the current available ELNs, their ease of use (or lack of it), and their accessibility issues across different devices and operating systems. Evidence suggests that whilst scientists willingly make use of generic notebooking software, spreadsheets, and other general office and scientific tools to aid their work, current ELNs are lacking in the required functionality to meet the needs of researchers. In this paper we present our extensive research and user study results, proposing an ELN built upon a pre-existing cloud notebook platform that makes use of accessible popular scientific software and semantic web technologies to help overcome the identified barriers to adoption. ('''[[Journal:Electronic lab notebooks: Can they replace paper|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 4–10:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab1 Kempler DataScienceJournal2017 16.png|240px]]</div> | |||
'''"[[Journal:Earth science data analytics: Definitions, techniques and skills|Earth science data analytics: Definitions, techniques and skills]]"''' | |||
The continuous evolution of data management systems affords great opportunities for the enhancement of knowledge and advancement of science research. To capitalize on these opportunities, it is essential to understand and develop methods that enable data relationships to be examined and [[information]] to be manipulated. Earth science data analytics (ESDA) comprises the techniques and skills needed to holistically extract information and knowledge from all sources of available, often heterogeneous, data sets. This paper reports on the ground-breaking efforts of the Earth Science Information Partners' (ESIP) ESDA Cluster in defining ESDA and identifying ESDA methodologies. As a result of the void of earth science data analytics in the literature, the ESIP ESDA definition and goals serve as an initial framework for a common understanding of techniques and skills that are available, as well as those still needed to support ESDA. Through the acquisition of earth science research use cases and categorization of ESDA result oriented research goals, ESDA techniques/skills have been assembled. The resulting ESDA techniques/skills provide the community with a definition for ESDA that is useful in articulating data management and research needs, as well as a working list of techniques and skills relevant to the different types of ESDA. ('''[[Journal:Earth science data analytics: Definitions, techniques and skills|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 28–September 3:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
'''"[[Journal:Bioinformatics: Indispensable, yet hidden in plain sight|Bioinformatics: Indispensable, yet hidden in plain sight]]"''' | |||
[[Bioinformatics]] has multitudinous identities, organizational alignments and disciplinary links. This variety allows bioinformaticians and bioinformatic work to contribute to much (if not most) of life science research in profound ways. The multitude of bioinformatic work also translates into a multitude of credit-distribution arrangements, apparently dismissing that work. | |||
We report on the epistemic and social arrangements that characterize the relationship between bioinformatics and life science. We describe, in sociological terms, the character, power and future of bioinformatic work. The character of bioinformatic work is such that its cultural, institutional and technical structures allow for it to be black-boxed easily. The result is that bioinformatic expertise and contributions travel easily and quickly, yet remain largely uncredited. The power of bioinformatic work is shaped by its dependency on life science work, which combined with the black-boxed character of bioinformatic expertise further contributes to situating bioinformatics on the periphery of the life sciences. Finally, the imagined futures of bioinformatic work suggest that bioinformatics will become ever more indispensable without necessarily becoming more visible, forcing bioinformaticians into difficult professional and career choices. ('''[[Journal:Global data quality assessment and the situated nature of “best” research practices in biology|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 21–27:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
'''"[[Journal:Global data quality assessment and the situated nature of “best” research practices in biology|Global data quality assessment and the situated nature of “best” research practices in biology]]"''' | |||
This paper reflects on the relation between international debates around data quality assessment and the diversity characterizing research practices, goals and environments within the life sciences. Since the emergence of molecular approaches, many biologists have focused their research, and related methods and instruments for data production, on the study of genes and genomes. While this trend is now shifting, prominent institutions and companies with stakes in molecular biology continue to set standards for what counts as "good science" worldwide, resulting in the use of specific data production technologies as proxy for assessing data quality. This is problematic considering (1) the variability in research cultures, goals and the very characteristics of biological systems, which can give rise to countless different approaches to knowledge production; and (2) the existence of research environments that produce high-quality, significant datasets despite not availing themselves of the latest technologies. Ethnographic research carried out in such environments evidences a widespread fear among researchers that providing extensive information about their experimental set-up will affect the perceived quality of their data, making their findings vulnerable to criticisms by better-resourced peers. These fears can make scientists resistant to sharing data or describing their provenance. ('''[[Journal:Global data quality assessment and the situated nature of “best” research practices in biology|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 14–20:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Grigis FInNeuroinformatics2017 11.jpg|240px]]</div> | |||
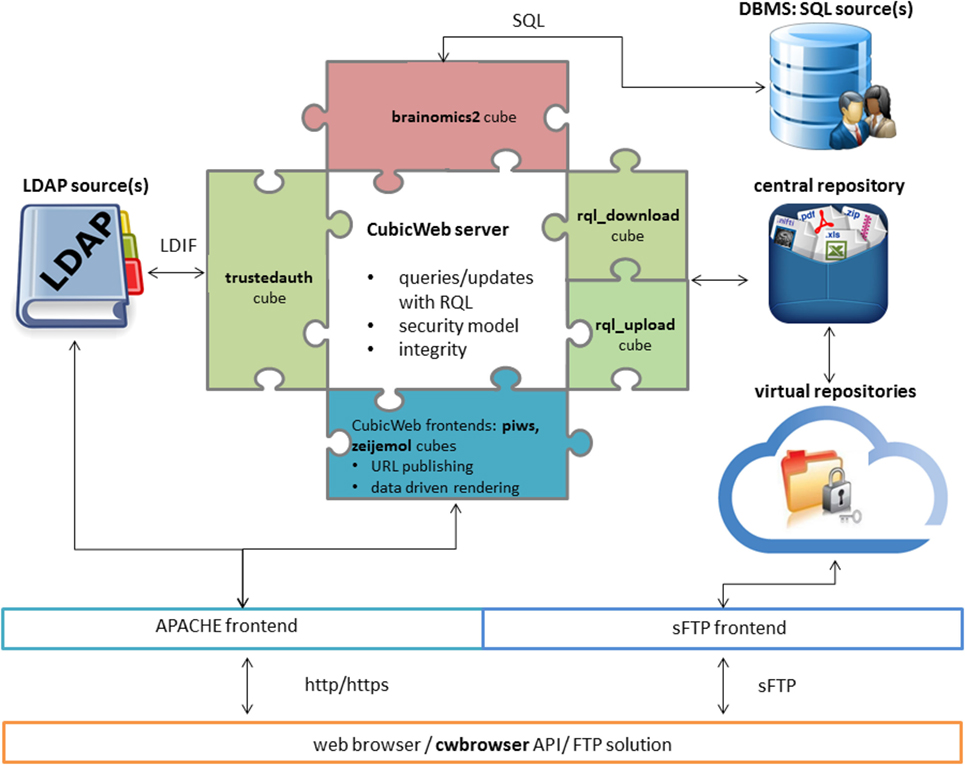
'''"[[Journal:Neuroimaging, genetics, and clinical data sharing in Python using the CubicWeb framework|Neuroimaging, genetics, and clinical data sharing in Python using the CubicWeb framework]]"''' | |||
In neurosciences or psychiatry, the emergence of large multi-center population imaging studies raises numerous technological challenges. From distributed data collection, across different institutions and countries, to final data publication service, one must handle the massive, heterogeneous, and complex data from genetics, imaging, demographics, or clinical scores. These data must be both efficiently obtained and downloadable. We present a Python solution, based on the CubicWeb open-source semantic framework, aimed at building population imaging study repositories. In addition, we focus on the tools developed around this framework to overcome the challenges associated with data sharing and collaborative requirements. We describe a set of three highly adaptive web services that transform the CubicWeb framework into a (1) multi-center upload platform, (2) collaborative quality assessment platform, and (3) publication platform endowed with massive-download capabilities. Two major European projects, IMAGEN and EU-AIMS, are currently supported by the described framework. We also present a Python package that enables end users to remotely query neuroimaging, genetics, and clinical data from scripts. ('''[[Journal:Neuroimaging, genetics, and clinical data sharing in Python using the CubicWeb framework|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 7–13:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Heo BMCBioinformatics2017 18.gif|240px]]</div> | |||
'''"[[Journal:Analyzing the field of bioinformatics with the multi-faceted topic modeling technique|Analyzing the field of bioinformatics with the multi-faceted topic modeling technique]]"''' | |||
[[Bioinformatics]] is an interdisciplinary field at the intersection of molecular biology and computing technology. To characterize the field as a convergent domain, researchers have used bibliometrics, augmented with text-mining techniques for content analysis. In previous studies, Latent Dirichlet Allocation (LDA) was the most representative topic modeling technique for identifying topic structure of subject areas. However, as opposed to revealing the topic structure in relation to metadata such as authors, publication date, and journals, LDA only displays the simple topic structure. | |||
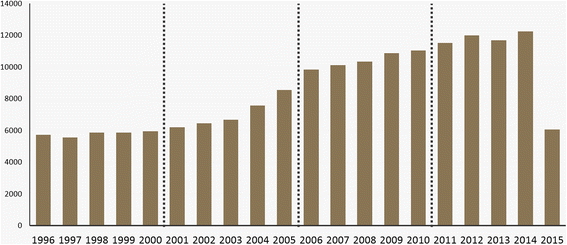
In this paper, we adopt the Author-Conference-Topic (ACT) model of Tang ''et al.'' to study the field of bioinformatics from the perspective of keyphrases, authors, and journals. The ACT model is capable of incorporating the paper, author, and conference into the topic distribution simultaneously. To obtain more meaningful results, we used journals and keyphrases instead of conferences and the bag-of-words. For analysis, we used PubMed to collect forty-six bioinformatics journals from the MEDLINE database. We conducted time series topic analysis over four periods from 1996 to 2015 to further examine the interdisciplinary nature of bioinformatics. ('''[[Journal:Analyzing the field of bioinformatics with the multi-faceted topic modeling technique|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 31–August 6:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Khan BMCBioinformatics2017 18.gif|240px]]</div> | |||
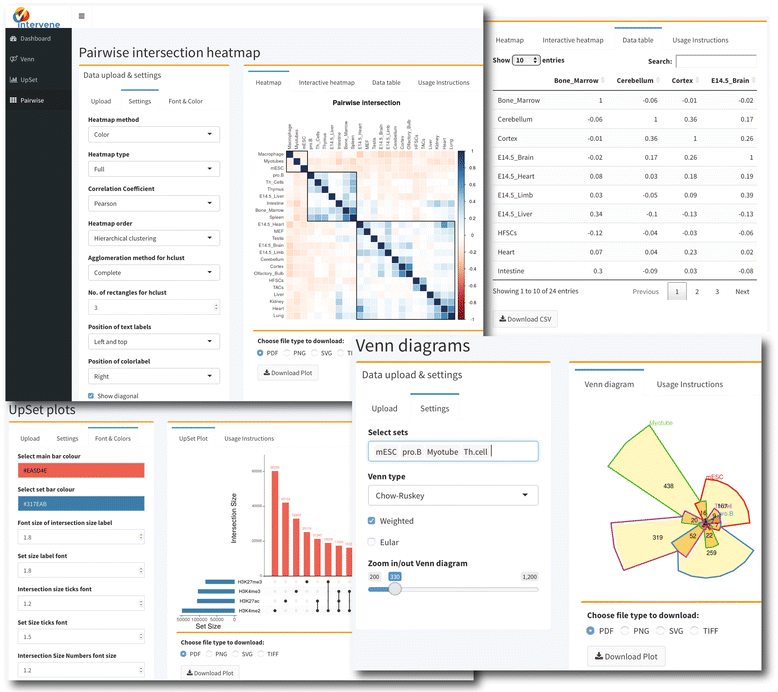
'''"[[Journal:Intervene: A tool for intersection and visualization of multiple gene or genomic region sets|Intervene: A tool for intersection and visualization of multiple gene or genomic region sets]]"''' | |||
A common task for scientists relies on comparing lists of genes or genomic regions derived from high-throughput sequencing experiments. While several tools exist to intersect and visualize sets of genes, similar tools dedicated to the visualization of genomic region sets are currently limited. | |||
To address this gap, we have developed the Intervene tool, which provides an easy and automated interface for the effective intersection and visualization of genomic region or list sets, thus facilitating their analysis and interpretation. Intervene contains three modules: ''venn'' to generate Venn diagrams of up to six sets, ''upset'' to generate UpSet plots of multiple sets, and ''pairwise'' to compute and visualize intersections of multiple sets as clustered heat maps. Intervene, and its interactive web ShinyApp companion, generate publication-quality figures for the interpretation of genomic region and list sets. ('''[[Journal:Intervene: A tool for intersection and visualization of multiple gene or genomic region sets|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 24–30:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
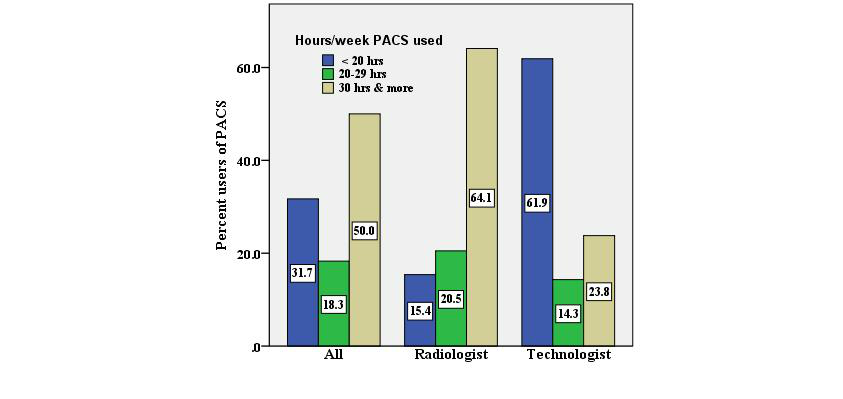
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Buabbas JMIRMedInfo2016 4-2.png|240px]]</div> | |||
'''"[[Journal:Users’ perspectives on a picture archiving and communication system (PACS): An in-depth study in a teaching hospital in Kuwait|Users’ perspectives on a picture archiving and communication system (PACS): An in-depth study in a teaching hospital in Kuwait]]"''' | |||
The [[picture archiving and communication system]] (PACS) is a well-known [[imaging informatics]] application in health care organizations, specifically designed for the radiology department. Health care providers have exhibited willingness toward evaluating PACS in hospitals to ascertain the critical success and failure of the technology, considering that evaluation is a basic requirement. | |||
This study aimed to evaluate the success of a PACS in a regional teaching hospital of Kuwait, from users’ perspectives, using information systems success criteria. | |||
An in-depth study was conducted by using quantitative and qualitative methods. This mixed-method study was based on: (1) questionnaires, distributed to all radiologists and technologists and (2) interviews, conducted with PACS administrators. ('''[[Journal:Users’ perspectives on a picture archiving and communication system (PACS): An in-depth study in a teaching hospital in Kuwait|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 17–23:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
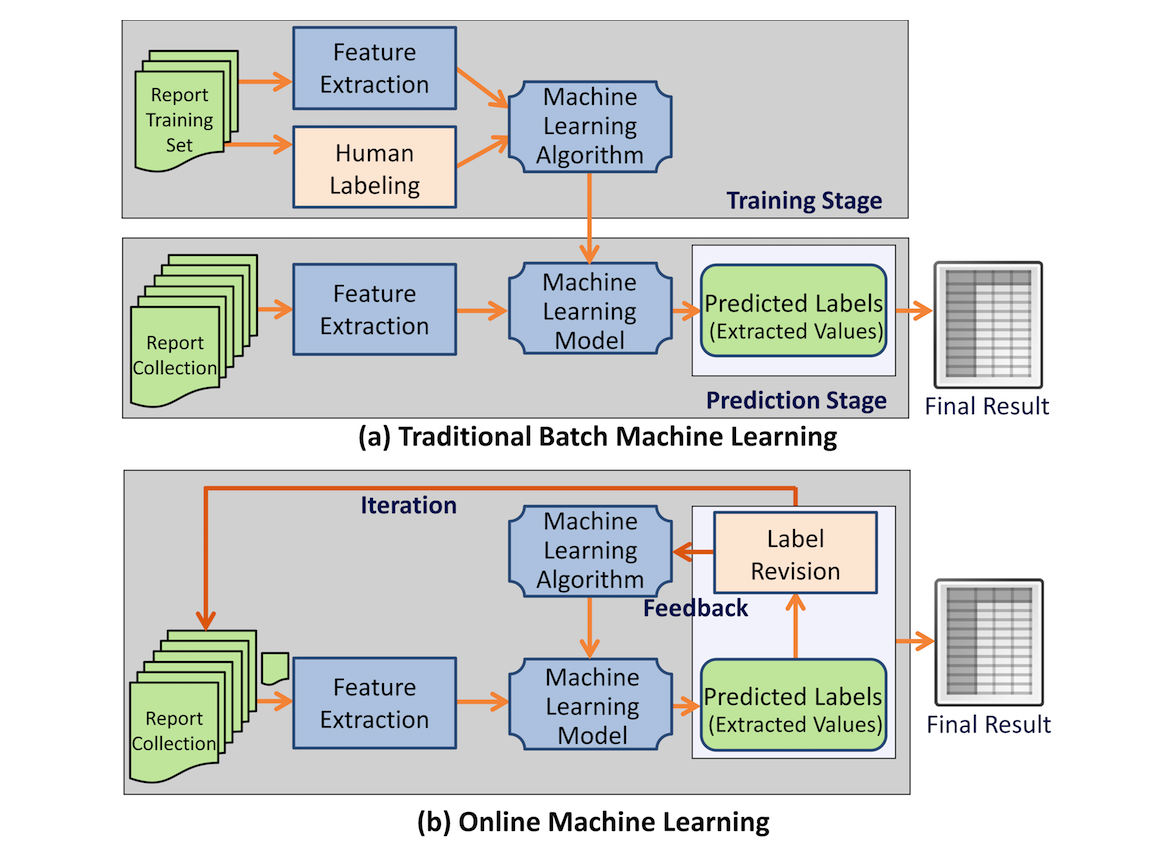
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Zheng JMIRMedInfo2017 5-2.png|240px]]</div> | |||
'''"[[Journal:Effective information extraction framework for heterogeneous clinical reports using online machine learning and controlled vocabularies|Effective information extraction framework for heterogeneous clinical reports using online machine learning and controlled vocabularies]]"''' | |||
Extracting structured data from narrated medical reports is challenged by the complexity of heterogeneous structures and vocabularies and often requires significant manual effort. Traditional machine-based approaches lack the capability to take user feedback for improving the extraction algorithm in real time. | |||
Our goal was to provide a generic [[information]] extraction framework that can support diverse clinical reports and enables a dynamic interaction between a human and a machine that produces highly accurate results. | |||
A clinical information extraction system IDEAL-X has been built on top of online machine learning. It processes one document at a time, and user interactions are recorded as feedback to update the learning model in real time. The updated model is used to predict values for extraction in subsequent documents. Once prediction accuracy reaches a user-acceptable threshold, the remaining documents may be batch processed. A customizable controlled vocabulary may be used to support extraction. ('''[[Journal:Effective information extraction framework for heterogeneous clinical reports using online machine learning and controlled vocabularies|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 10–16:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab1 Kyobe BiopresBiobank2017 15-2.png|240px]]</div> | |||
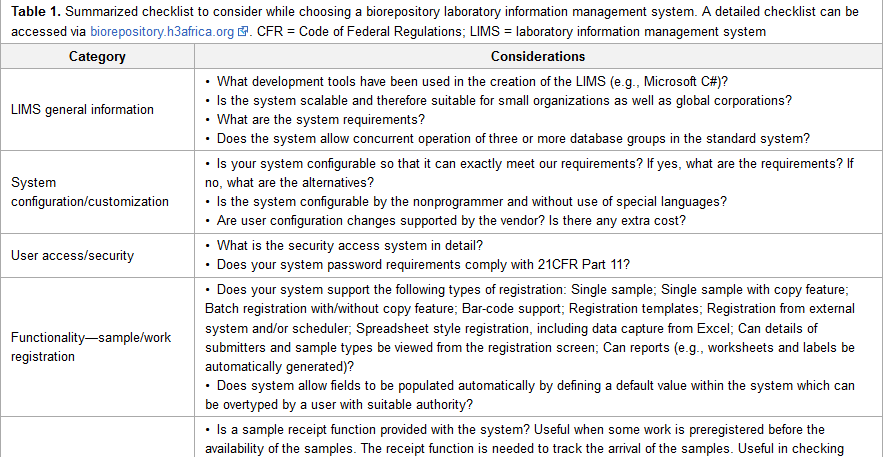
'''"[[Journal:Selecting a laboratory information management system for biorepositories in low- and middle-income countries: The H3Africa experience and lessons learned|Selecting a laboratory information management system for biorepositories in low- and middle-income countries: The H3Africa experience and lessons learned]]"''' | |||
Biorepositories in Africa need significant infrastructural support to meet International Society for Biological and Environmental Repositories (ISBER) Best Practices to support population-based genomics research. ISBER recommends a biorepository information management system which can manage workflows from biospecimen receipt to distribution. The H3Africa Initiative set out to develop regional African biorepositories where Uganda, Nigeria, and South Africa were successfully awarded grants to develop the state-of-the-art biorepositories. The biorepositories carried out an elaborate process to evaluate and choose a [[laboratory information management system]] (LIMS) with the aim of integrating the three geographically distinct sites. In this article, we review the processes, African experience, and lessons learned, and we make recommendations for choosing a biorepository LIMS in the African context. ('''[[Selecting a laboratory information management system for biorepositories in low- and middle-income countries: The H3Africa experience and lessons learned|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 3–9:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Bendou BiopresAndBiobank2017 15-2.gif|240px]]</div> | |||
'''"[[Journal:Baobab Laboratory Information Management System: Development of an open-source laboratory information management system for biobanking|Baobab Laboratory Information Management System: Development of an open-source laboratory information management system for biobanking]]"''' | |||
A [[laboratory information management system]] (LIMS) is central to the [[informatics]] infrastructure that underlies [[biobanking]] activities. To date, a wide range of commercial and open-source LIMSs are available, and the decision to opt for one LIMS over another is often influenced by the needs of the biobank clients and researchers, as well as available financial resources. The Baobab LIMS was developed by customizing the [[Bika LIMS]] software to meet the requirements of biobanking best practices. The need to implement biobank standard operation procedures as well as stimulate the use of standards for biobank data representation motivated the implementation of Baobab LIMS, an open-source LIMS for biobanking. Baobab LIMS comprises modules for biospecimen kit assembly, shipping of biospecimen kits, storage management, analysis requests, reporting, and invoicing. The Baobab LIMS is based on the Plone web-content management framework. All the system requirements for Plone are applicable to Baobab LIMS, including the need for a server with at least 8 GB RAM and 120 GB hard disk space. Baobab LIMS is a client-server-based system, whereby the end user is able to access the system securely through the internet on a standard web browser, thereby eliminating the need for standalone installations on all machines. ('''[[Baobab Laboratory Information Management System: Development of an open-source laboratory information management system for biobanking|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 19–25:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;"><div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 D'Anca NatHazEarth2017 17-2.png|240px]]</div> | |||
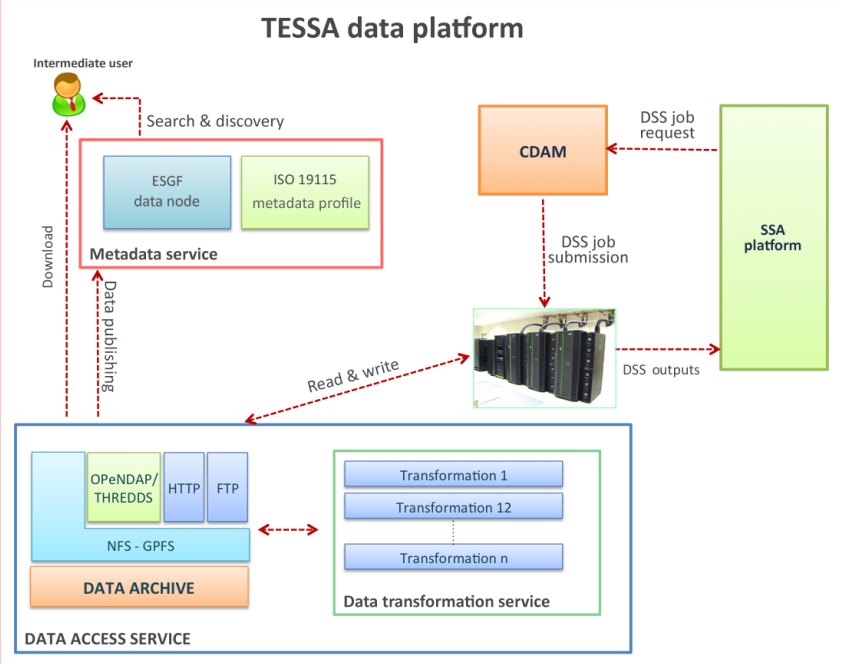
'''"[[Journal:A multi-service data management platform for scientific oceanographic products|A multi-service data management platform for scientific oceanographic products]]"''' | |||
An efficient, secure and interoperable data platform solution has been developed in the TESSA project to provide fast navigation and access to the data stored in the data archive, as well as a standard-based metadata management support. The platform mainly targets scientific users and the situational sea awareness high-level services such as the decision support systems (DSS). These datasets are accessible through the following three main components: the Data Access Service (DAS), the Metadata Service and the Complex Data Analysis Module (CDAM). The DAS allows access to data stored in the archive by providing interfaces for different protocols and services for downloading, variable selection, data subsetting or map generation. Metadata Service is the heart of the information system of TESSA products and completes the overall infrastructure for data and metadata management. This component enables data search and discovery and addresses interoperability by exploiting widely adopted standards for geospatial data. Finally, the CDAM represents the back end of the TESSA DSS by performing on-demand complex data analysis tasks. ('''[[A multi-service data management platform for scientific oceanographic products|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 26–July 2:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
'''"[[Journal:The FAIR Guiding Principles for scientific data management and stewardship|The FAIR Guiding Principles for scientific data management and stewardship]]"''' | |||
There is an urgent need to improve the infrastructure supporting the reuse of scholarly data. A diverse set of stakeholders — representing academia, industry, funding agencies, and scholarly publishers — have come together to design and jointly endorse a concise and measureable set of principles that we refer to as the FAIR Data Principles. The intent is that these may act as a guideline for those wishing to enhance the reusability of their data holdings. Distinct from peer initiatives that focus on the human scholar, the FAIR Principles put specific emphasis on enhancing the ability of machines to automatically find and use the data, in addition to supporting its reuse by individuals. This comment article represents the first formal publication of the FAIR Principles, and it includes the rationale behind them as well as some exemplar implementations in the community. ('''[[The FAIR Guiding Principles for scientific data management and stewardship|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 19–25:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;"><div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 D'Anca NatHazEarth2017 17-2.png|240px]]</div> | |||
'''"[[Journal:A multi-service data management platform for scientific oceanographic products|A multi-service data management platform for scientific oceanographic products]]"''' | |||
An efficient, secure and interoperable data platform solution has been developed in the TESSA project to provide fast navigation and access to the data stored in the data archive, as well as a standard-based metadata management support. The platform mainly targets scientific users and the situational sea awareness high-level services such as the decision support systems (DSS). These datasets are accessible through the following three main components: the Data Access Service (DAS), the Metadata Service and the Complex Data Analysis Module (CDAM). The DAS allows access to data stored in the archive by providing interfaces for different protocols and services for downloading, variable selection, data subsetting or map generation. Metadata Service is the heart of the information system of TESSA products and completes the overall infrastructure for data and metadata management. This component enables data search and discovery and addresses interoperability by exploiting widely adopted standards for geospatial data. Finally, the CDAM represents the back end of the TESSA DSS by performing on-demand complex data analysis tasks. ('''[[A multi-service data management platform for scientific oceanographic products|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 12–18:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;"><div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Hunter Metabolomics2017 13-2.gif|80px]]</div> | |||
'''"[[Journal:MASTR-MS: A web-based collaborative laboratory information management system (LIMS) for metabolomics|MASTR-MS: A web-based collaborative laboratory information management system (LIMS) for metabolomics]]"''' | |||
An increasing number of research [[laboratory|laboratories]] and core analytical facilities around the world are developing high throughput metabolomic analytical and data processing pipelines that are capable of handling hundreds to thousands of individual samples per year, often over multiple projects, collaborations and sample types. At present, there are no [[laboratory information management system]]s (LIMS) that are specifically tailored for metabolomics laboratories that are capable of tracking samples and associated metadata from the beginning to the end of an experiment, including data processing and archiving, and which are also suitable for use in large institutional core facilities or multi-laboratory consortia as well as single laboratory environments. | |||
Here we present [[MASTR-MS]], a downloadable and installable LIMS solution that can be deployed either within a single laboratory or used to link workflows across a multisite network.('''[[MASTR-MS: A web-based collaborative laboratory information management system (LIMS) for metabolomics|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 5–11:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;"><div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Martins BMCMedInfoDecMak2017 17-1.gif|240px]]</div> | |||
'''"[[Journal:The effect of a test ordering software intervention on the prescription of unnecessary laboratory tests - A randomized controlled trial|The effect of a test ordering software intervention on the prescription of unnecessary laboratory tests - A randomized controlled trial]]"''' | |||
The way [[electronic health record]] and [[Computerized physician order entry|laboratory test ordering system]] software is designed may influence physicians’ prescription. A randomized controlled trial was performed to measure the impact of a diagnostic and laboratory tests ordering system software modification. | |||
Participants were family physicians working and prescribing diagnostic and [[laboratory]] tests. The intervention group had modified software with basic shortcut menu changes, where some tests were withdrawn or added, and with the implementation of an evidence-based [[clinical decision support system]] based on United States Preventive Services Task Force (USPSTF) recommendations. This intervention group was compared with typically used software (control group). | |||
The outcomes were the number of tests prescribed from those: withdrawn from the basic menu; added to the basic menu; marked with green dots (USPSTF’s grade A and B); and marked with red dots (USPSTF’s grade D). ('''[[The effect of a test ordering software intervention on the prescription of unnecessary laboratory tests - A randomized controlled trial|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 29–June 4:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;"><div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab2 Alsaffar JMIRMedicalInfo2017 5-1.png|240px]]</div> | |||
'''"[[Journal:The state of open-source electronic health record projects: A software anthropology study|The state of open-source electronic health record projects: A software anthropology study]]"''' | |||
[[Electronic health record]]s (EHR) are a key tool in managing and storing patients’ [[information]]. Currently, there are over 50 open-source EHR systems available. Functionality and usability are important factors for determining the success of any system. These factors are often a direct reflection of the domain knowledge and developers’ motivations. However, few published studies have focused on the characteristics of [[free and open-source software]] (F/OSS) EHR systems, and none to date have discussed the motivation, knowledge background, and demographic characteristics of the developers involved in open-source EHR projects. | |||
This study analyzed the characteristics of prevailing F/OSS EHR systems and aimed to provide an understanding of the motivation, knowledge background, and characteristics of the developers. ('''[[The state of open-source electronic health record projects: A software anthropology study|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 22–28:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;"><div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Eyal-Altman BMCBioinformatics2017 18.gif|240px]]</div> | |||
'''"[[Journal:PCM-SABRE: A platform for benchmarking and comparing outcome prediction methods in precision cancer medicine|PCM-SABRE: A platform for benchmarking and comparing outcome prediction methods in precision cancer medicine]]"''' | |||
Numerous publications attempt to predict cancer survival outcome from gene expression data using machine-learning methods. A direct comparison of these works is challenging for the following reasons: (1) inconsistent measures used to evaluate the performance of different models, and (2) incomplete specification of critical stages in the process of knowledge discovery. There is a need for a platform that would allow researchers to replicate previous works and to test the impact of changes in the knowledge discovery process on the accuracy of the induced models. | |||
We developed the PCM-SABRE platform, which supports the entire knowledge discovery process for cancer outcome analysis. PCM-SABRE was developed using [[KNIME]]. By using PCM-SABRE to reproduce the results of previously published works on breast cancer survival, we define a baseline for evaluating future attempts to predict cancer outcome with machine learning. ('''[[PCM-SABRE: A platform for benchmarking and comparing outcome prediction methods in precision cancer medicine|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 15–21:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
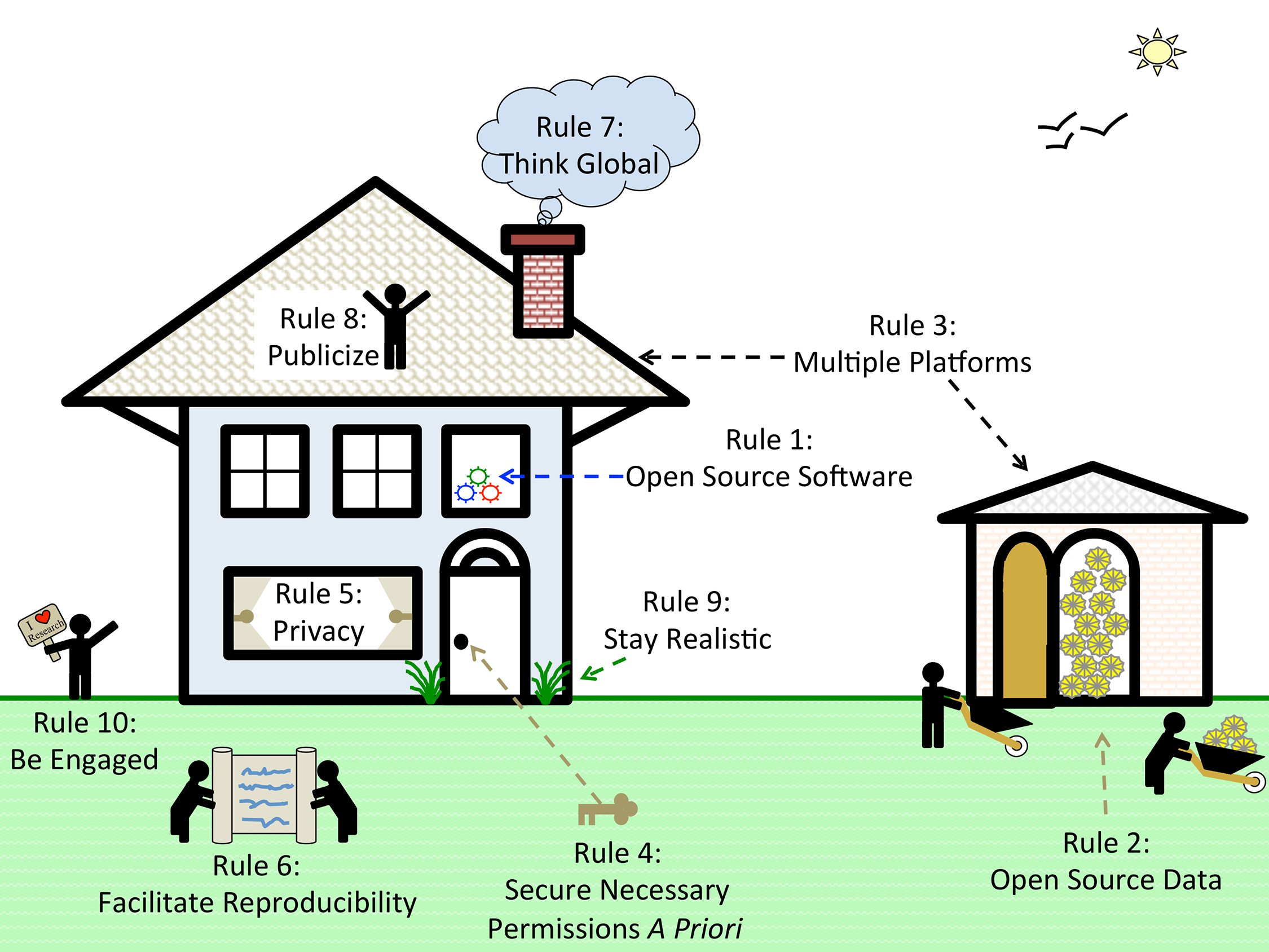
'''"[[Journal:Ten simple rules for cultivating open science and collaborative R&D|Ten simple rules for cultivating open science and collaborative R&D]]"''' | |||
How can we address the complexity and cost of applying science to societal challenges? | |||
Open science and collaborative R&D may help. Open science has been described as "a research accelerator." Open science implies open access but goes beyond it: "Imagine a connected online web of scientific knowledge that integrates and connects data, computer code, chains of scientific reasoning, descriptions of open problems, and beyond ... tightly integrated with a scientific social web that directs scientists' attention where it is most valuable, releasing enormous collaborative potential." | |||
Open science and collaborative approaches are often described as open-source, by analogy with open-source software such as the operating system Linux which powers Google and Amazon — collaboratively created software which is free to use and adapt, and popular for internet infrastructure and scientific research. ('''[[Journal:Ten simple rules for cultivating open science and collaborative R&D|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 8–14:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"><div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Boland PLOSCompBio2017 13-1.png|240px]]</div> | |||
'''"[[Journal:Ten simple rules to enable multi-site collaborations through data sharing|Ten simple rules to enable multi-site collaborations through data sharing]]"''' | |||
Open access, open data, and software are critical for advancing science and enabling collaboration across multiple institutions and throughout the world. Despite near universal recognition of its importance, major barriers still exist to sharing raw data, software, and research products throughout the scientific community. Many of these barriers vary by specialty, increasing the difficulties for interdisciplinary and/or translational researchers to engage in collaborative research. Multi-site collaborations are vital for increasing both the impact and the generalizability of research results. However, they often present unique data sharing challenges. We discuss enabling multi-site collaborations through enhanced data sharing in this set of ''Ten Simple Rules''. ('''[[Journal:Ten simple rules to enable multi-site collaborations through data sharing|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 1–7:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
'''"[[Journal:Ten simple rules for developing usable software in computational biology|Ten simple rules for developing usable software in computational biology]]"''' | |||
The rise of high-throughput technologies in molecular biology has led to a massive amount of publicly available data. While computational method development has been a cornerstone of biomedical research for decades, the rapid technological progress in the wet [[laboratory]] makes it difficult for software development to keep pace. Wet lab scientists rely heavily on computational methods, especially since more research is now performed ''in silico''. However, suitable tools do not always exist, and not everyone has the skills to write complex software. Computational biologists are required to close this gap, but they often lack formal training in software engineering. To alleviate this, several related challenges have been previously addressed in the ''Ten Simple Rules'' series, including reproducibility, effectiveness, and open-source development of software. ('''[[Journal:Ten simple rules for developing usable software in computational biology|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 24–30:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
'''"[[Journal:The effect of the General Data Protection Regulation on medical research|The effect of the General Data Protection Regulation on medical research]]"''' | |||
The enactment of the General Data Protection Regulation (GDPR) will impact on European data science. Particular concerns relating to consent requirements that would severely restrict medical data research have been raised. Our objective is to explain the changes in data protection laws that apply to medical research and to discuss their potential impact ... The GDPR makes the classification of pseudonymised data as personal data clearer, although it has not been entirely resolved. Biomedical research on personal data where consent has not been obtained must be of substantial public interest. [We conclude] [t]he GDPR introduces protections for data subjects that aim for consistency across the E.U. The proposed changes will make little impact on biomedical data research. ('''[[Journal:The effect of the General Data Protection Regulation on medical research|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 17–23:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Rubel FInNeuroinformatics2016 10.jpg|240px]]</div> | |||
'''"[[Journal:Methods for specifying scientific data standards and modeling relationships with applications to neuroscience|Methods for specifying scientific data standards and modeling relationships with applications to neuroscience]]"''' | |||
Neuroscience continues to experience a tremendous growth in data; in terms of the volume and variety of data, the velocity at which data is acquired, and in turn the veracity of data. These challenges are a serious impediment to sharing of data, analyses, and tools within and across labs. Here, we introduce BRAINformat, a novel data standardization framework for the design and management of scientific data formats. The BRAINformat library defines application-independent design concepts and modules that together create a general framework for standardization of scientific data. We describe the formal specification of scientific data standards, which facilitates sharing and verification of data and formats. We introduce the concept of "managed objects," enabling semantic components of data formats to be specified as self-contained units, supporting modular and reusable design of data format components and file storage. We also introduce the novel concept of "relationship attributes" for modeling and use of semantic relationships between data objects. Based on these concepts we demonstrate the application of our framework to design and implement a standard format for electrophysiology data and show how data standardization and relationship-modeling facilitate [[data analysis]] and sharing. ('''[[Journal:Methods for specifying scientific data standards and modeling relationships with applications to neuroscience|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 10–16:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Khalsa DataScienceJ2017 16-1.png|240px]]</div> | |||
'''"[[Journal:Data and metadata brokering – Theory and practice from the BCube Project|Data and metadata brokering – Theory and practice from the BCube Project]]"''' | |||
EarthCube is a U.S. National Science Foundation initiative that aims to create a cyberinfrastructure (CI) for all the geosciences. An initial set of "building blocks" was funded to develop potential components of that CI. The Brokering Building Block (BCube) created a brokering framework to demonstrate cross-disciplinary data access based on a set of use cases developed by scientists from the domains of hydrology, oceanography, polar science and climate/weather. While some successes were achieved, considerable challenges were encountered. We present a synopsis of the processes and outcomes of the BCube experiment. ('''[[Journal:Data and metadata brokering – Theory and practice from the BCube Project|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 3–9:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | <div style="padding:0.4em 1em 0.3em 1em;"> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Harvey JoCheminformatics2017 9.gif|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Harvey JoCheminformatics2017 9.gif|240px]]</div> | ||
Latest revision as of 19:57, 3 April 2024
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 - 2023 - 2024 |
Featured article of the week archive - 2017
Welcome to the LIMSwiki 2017 archive for the Featured Article of the Week.
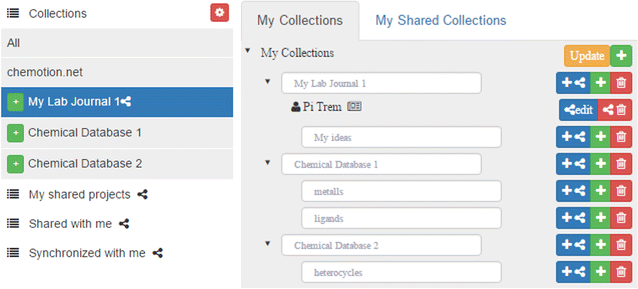
Featured article of the week: December 25–31:"Chemotion ELN: An open-source electronic lab notebook for chemists in academia" The development of an electronic laboratory notebook (ELN) for researchers working in the field of chemical sciences is presented. The web-based application is available as open-source software that offers modern solutions for chemical researchers. The Chemotion ELN is equipped with the basic functionalities necessary for the acquisition and processing of chemical data, in particular work with molecular structures and calculations based on molecular properties. The ELN supports planning, description, storage, and management for the routine work of organic chemists. It also provides tools for communicating and sharing the recorded research data among colleagues. Meeting the requirements of a state-of-the-art research infrastructure, the ELN allows the search for molecules and reactions not only within the user’s data but also in conventional external sources as provided by SciFinder and PubChem. The presented development makes allowance for the growing dependency of scientific activity on the availability of digital information by providing open- source instruments to record and reuse research data. The current version of the ELN has been used for over half of a year in our chemistry research group, serving as a common infrastructure for chemistry research and enabling chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms. (Full article...)
|