Journal:Analyzing the field of bioinformatics with the multi-faceted topic modeling technique
| Full article title | Analyzing the field of bioinformatics with the multi-faceted topic modeling technique |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) | Heo, Go Eun; Kang, Keun Young; Song, Min; Lee, Jeong-Hoon |
| Author affiliation(s) | Yonsei University, POSTECH |
| Primary contact | Email: min dot song at yonsei dot ac dot kr |
| Year published | 2017 |
| Volume and issue | 18 (Suppl 7) |
| Page(s) | 251 |
| DOI | 10.1186/s12859-017-1640-x |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1640-x |
| Download | https://bmcbioinformatics.biomedcentral.com/track/pdf/10.1186/s12859-017-1640-x (PDF) |
Abstract
Background: Bioinformatics is an interdisciplinary field at the intersection of molecular biology and computing technology. To characterize the field as a convergent domain, researchers have used bibliometrics, augmented with text-mining techniques for content analysis. In previous studies, Latent Dirichlet Allocation (LDA) was the most representative topic modeling technique for identifying topic structure of subject areas. However, as opposed to revealing the topic structure in relation to metadata such as authors, publication date, and journals, LDA only displays the simple topic structure.
Methods: In this paper, we adopt the Author-Conference-Topic (ACT) model of Tang et al. to study the field of bioinformatics from the perspective of keyphrases, authors, and journals. The ACT model is capable of incorporating the paper, author, and conference into the topic distribution simultaneously. To obtain more meaningful results, we used journals and keyphrases instead of conferences and the bag-of-words. For analysis, we used PubMed to collect forty-six bioinformatics journals from the MEDLINE database. We conducted time series topic analysis over four periods from 1996 to 2015 to further examine the interdisciplinary nature of bioinformatics.
Results: We analyzed the ACT Model results in each period. Additionally, for further integrated analysis, we conducted a time series analysis among the top-ranked keyphrases, journals, and authors according to their frequency. We also examined the patterns in the top journals by simultaneously identifying the topical probability in each period, as well as the top authors and keyphrases. The results indicate that in recent years diversified topics have become more prevalent, and convergent topics have become more clearly represented.
Conclusion: The results of our analysis imply that over time the field of bioinformatics becomes more interdisciplinary where there is a steady increase in peripheral fields such as conceptual, mathematical, and system biology. These results are confirmed by integrated analysis of topic distribution as well as top ranked keyphrases, authors, and journals.
Keywords: bioinformatics, text mining, topic modeling, ACT model, keyphrase extraction
Background
Over the years, academic subject areas have converged to form a variety of new, interdisciplinary fields. Bioinformatics is one example. Research domains from molecular biology to machine learning are used in conjunction to better understand complex biological systems such as cells, tissues, and the human body. Due to the complexity and broadness of the field, bibliometric analysis is often adopted to assess the current knowledge structure of a subject area, specify the current research themes, and identify the core literature of that area.[1]
Bibliometrics identifies research trends using quantitative measures such as a researcher’s number of publications and citations, journal impact factors, and other indices that can measure impact or productivity of author or journal.[2][3][4][5] In addition, other factors such as the affiliation of authors, collaborations, and citation data are often incorporated into bibliometric analysis.[6][7][8][9]
Previous studies mainly rely on quantitative measures and suffer from the lack of content analysis. To incorporate content analysis into bibliometrics, text-mining techniques are applied. Topic-modeling techniques are mostly adopted to identify the topics of a subject area while analyzing that area more abundantly.[10][11][12][13] These techniques allow for enriched content analysis. As an extension of Latent Dirichlet Allocation (LDA), which is the best received topic-modeling technique, Steyvers et al.[14] proposed the author-topic modeling technique that analyzes authors and topics simultaneously. They identify the authors’ impact or productivity of researchers in a given subject area.[15][16] By adding multiple conditions to LDA, Tang et al.[17] suggested a new methodology, called the Author-Conference-Topic (ACT) model that analyzes the author, conference, and topic in one model to understand the subject area in an integrated manner.
In this paper, we apply the ACT model to examine the interdisciplinary nature of bioinformatics. Unlike studies that use extended versions of LDA for topic analysis, the ACT model enables us to analyze topic, author, and journal at one time, providing an integrated view for understanding bioinformatics. The research questions that we are to investigate in this paper are: 1) What are the topical trends of bioinformatics over time? 2) Who are the key contributors in major topics of bioinformatics?, and 3) Which journal is leading which topic?
To address these questions, we collect PubMed articles in XML format and extract metadata and content such as the PMID, author, year, journal, title, and abstract. From the title and abstract, we extract keyphrases, which provide more meaningful interpretations than single words, as an input of the ACT model. We also divide the collected datasets into four time periods to examine the topic changes over time. The results of ACT model–based analysis show that various topics begin to appear and mixed subject topics become more apparent over time.
The rest of the paper is organized as follows. In the "Related work" section, we discuss work related to bibliometric analysis and topic modeling. We then describe the proposed method in the "Methods" section. We analyze and discuss the results of leading topics, authors, and journals in the "Results" and "Discussion" sections. Finally, we conclude the paper and suggest future lines of inquiry in the "Conclusion" section.
Related work
Bibliometric analysis
Bibliometric analysis identifies the research trends in a given subject area and core journals or documents, and it helps with contrasting analysis. Many bibliometric studies use the number of published articles or journal impact factors to measure research productivity or to identify core journals in a specific field. Soteriades and Falalgas[3] applied quantitative and qualitative measurements to analyze the fields of preventive medicine, occupational and environmental medicine, epidemiology, and public health using the number of articles and impact factor. Ugolini et al.[4] measured research productivity and evaluated the publication trends in the field of cancer molecular epidemiology. To quantify productivity, they used the number of articles and the average and sum of impact factors. To evaluate publication trends, they collected and divided the keywords from MeSH terms about the publication into six groups. Ramos et al.[18] measured the national research activity of the tuberculosis field, using impact factor and the first author’s address. Claude et al.[19] examined research productivity by using distribution of publications related to medicine and ANN, the subfield of biology. They used the number of publications, impact factor, and journal category compared with national gross domestic product (GDP). In the bioinformatics field, Patra and Mishra[20] used the number of articles, publication of each journal, publication type, and the impact factor of journals to understand the growth of bioinformatics. They also found the core journals in the bioinformatics fields. Using author affiliation, they applied Lotka’s law to assess the distribution of each author’s productivity. Chen et al.[2] identified research trends using statistical methods based on the type of publication, language, and distribution of nation or institution. They measured h-index, adding statistical materials with the number of citations. Through this, they analyzed the research productivity by topic, institution, and journal. In addition, they conducted a keyword analysis to comprehend the research trend in a macroscopic view.
Mainstream bibliometrics research focuses on identifying the knowledge structure of a certain field with quantitative measures. In addition, some studies use author information or the collaboration pattern among authors to understand the certain field. Seglen and Aksnes[9] used the size and the productivity of research groups in the microbiology field in Norway as a measurement for bibliometric analysis. Geaney et al.[7] performed bibliometric analysis and density-equalizing mapping on scientific publications related to type 2 diabetes mellitus. They collected citation data and used various citation-oriented measures such as the number of citations, the average number of citations per journal, the total number of publications, impact factor, and Eigenfactor score. To conduct content analysis and study the collaboration pattern between authors and the core sub-field of AIDS, Macías-Chapula and Mijangos-Nolasco[8] analyzed MeSH thesaurus using check tags, main headings, and subheadings of each MeSH term hierarchy. In addition, to measure the national research productivity, they used the authors’ address information. Bornmann and Mutz[6] recently identified the development of modern science by bibliometric analysis. They divide the data into three time periods to analyze the changes of fields over time.
Text mining applied to bibliometrics
Recently, there have been many attempts to apply text-mining techniques to bibliometric analysis to identify the knowledge structure of the field or measure its influence on other researchers and their fields and productivity. Song and Kim[11] collected full-text articles from PubMed Central and computed their citation relation. They infer the knowledge structure and understand the trend of the bioinformatics field. In a similar vein, Song et al.[12] measured the influence and productivity of bioinformatics by mining full-text articles retrieved from PubMed Central. To calculate the field’s productivity, they identified the most productive author, nation, institution, and topic word; to calculate its influence, they identified the most-cited paper, author, and rising researcher. Song et al.[21] analyzed topic evolution in the bioinformatics field using DBLP data in the field of computer science. To identify topic trends over time, they divided a dozen years (2000–2011) into four periods and applied Markov Random Field-based topic clustering. For automatic clustering labeling, they calculated topic similarity based on Within-Period Cluster Similarity (WPCS) and Between-Period Cluster Similarity (BPCS). Their approach created topic graphs that show interaction among topics over some period of time. Lee et al.[22] mapped the Alzheimer’s disease field into three different perspectives: indexer, author, and citer. They applied entity-metrics[23] the extended notion of bibliometrics, to analyze the field by constructing four kinds of networks that convey these three perspectives.
These studies identify the knowledge structure of a certain field by constructing bibliometric networks or databases with text-mining techniques. The most prevalent approach is to apply topic modeling to content analysis as a part of bibliometrics. Starting from the probabilistic Latent Semantic Indexing (pLSI)[24] model, Latent Dirichlet Allocation (LDA)[25] is the most accepted topic modeling technique for bibliometrics. While each document consists of a set of topics in pLSI, using the LDA model results in a more precise manipulation being added to organize the topics. Yan[13] used the LDA model to measure the influence and popularity of library and information science. He also identified the most-cited area and the patterns in this field. Jeong and Song’s[10] research measured the time gap among three different resources — web, patent, and scientific publication — in two research domains by applying the LDA model. The basic input unit for LDA is a set of documents. To organize author information into topics, Rosen-Zvi et al.[16] and Steyvers et al.[14] proposed the author-topic model with different theoretical background. Li et al.[15] identified the relations between authors and topics by using the author-topic model. They analyzed the topic distribution to examine how many authors are associated with a certain topic. Also through the number of authors, they identified topics that are studied by many researchers. Tang et al.[17] proposed the ACT model which identifies paper, author, and conference simultaneously. Additionally, they developed the ArnetMiner system for mining academic research social networks. Tang et al.[26] also supplement ArnetMiner for a topic level expertise search over heterogeneous networks using the ACT model. It generates the most issued topics, author’s interestedness, paper search, academic suggestion, and experts in a specific field. Kim et al.[27] adopted the ACT model in terms of citation analysis. They collected their dataset in the field of oncology from PubMed Central, which provides the full-text articles in the biomedical field. They utilized the ACT model for analyzing citation sentences and journals instead of abstracts and conferences.
In conclusion, most previous studies identified knowledge structures by adopting not only bibliometric analysis but text-mining techniques such as the LDA model. To supplement bibliometric analysis, there are many attempts to incorporate content analysis into bibliometrics by adopting the LDA model text-mining techniques. However, the main limitation of this application of the LDA model, the representative method for trend analysis, is that it only explains topical trends by using one parameter such as the bag-of-words on documents via topical terms. It is not sufficient to conduct comprehensive analysis for understanding knowledge disciplines. Therefore, in this paper, we apply the ACT model to the bioinformatics field for integrated analysis. Applying the ACT model, we aim to explore the importance of authors and journals in relation to topics. We divided the collected datasets into four periods to trace the changes of topic, author, and journal ranking over time, combining the results with bibliometric analysis.
Methods
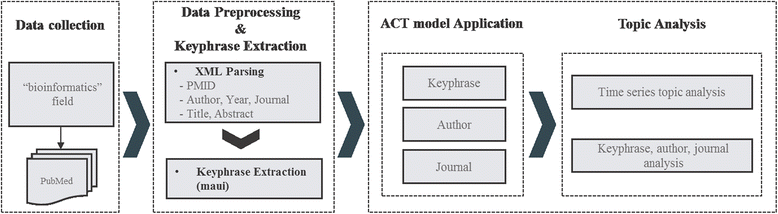
In this section, we describe data collection, preprocessing, and keyphrase extraction to feed input into the ACT model. Figure 1 illustrates the overflow of our approach; detailed descriptions of each component are provided in the following section.
|
Data collection
For analysis, we collected the 48 journals in the bioinformatics field as used by Song and Kim.[11] Forty-six out of the 48 journals were found via the advanced search tool provided by PubMed. Two journals, Advanced Bioinformatics and Genome Integration, were not retrieved from PubMed. We downloaded the 46 PubMed-listed journals in XML format (Table 1). The total number of papers indexed in these journals was 241,569; Biochemistry had the greatest number of papers with 62,270, accounting for 25.78% of the collected publications.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Data preprocessing and keyphrase extraction
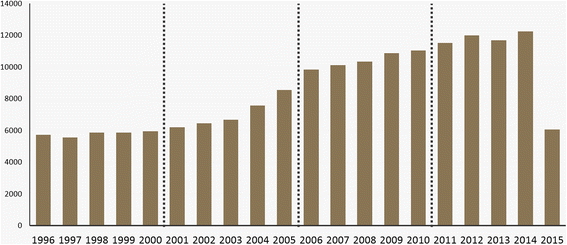
We limited the publication year back to 1996 and divided the dataset into the following four time periods to identify the trend of bioinformatics from the birth of the field to present: 1996–2000, 2001–2005, 2006–2010, and 2011–2015 (Fig. 2).
|
As shown in Fig. 2, there is a relatively consistent increase in the number of papers. There are fewer than half as many papers published in 2015 than in 2014 because we collected our dataset in June 2015. Nevertheless, we included the 2015 data to observe the latest publication trends.
Table 2 presents the breakdown of our dataset by period. As in Fig. 2, the fourth period is the most productive, containing 53,520 papers, or 31.46% of the total dataset. The most productive year is 2014, which accounts for 7.20% with 12,251 papers. The total number of papers for all 20 years is 170,099. This number is different from Table 1 (241,569) as a result of preprocessing; we excluded papers that did not have an abstract.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
We extract various metadata, such as the PMID, author, publication year, journal title, title, and abstract from XML formatted records. After XML processing, we combine the title with abstract and conduct keyphrase extraction. For keyphrase extraction, we use MAUI, which has the keyphrase model trained with MeSH terms.[28] In this dataset, there are 500 documents and several keys consisting of MeSH terms about each document, manually assigned by the indexer. MAUI is a newer version of the keyphrase extraction algorithm KEA.[29] Keyphrase extraction enables researchers to select representative phrases to make topic detection more meaningful. Therefore, we use keyphrases extracted from the title or abstract as our input for the ACT model instead of individual words.
Table 3 shows the results of keyphrase extraction and other metadata such as the title and publication year from the PubMed record PMID 26030820.
| ||||||||||||||
ACT model application
The ACT model, proposed by Tang et al.[17] as an extension of the LDA model[25], is a unified topic model for modeling various metadata simultaneously. This model starts with the assumption that the order of the topic created by the paper, author, and conference is the same. It also estimates the statistical distribution associated with all topics for the purpose of discovering latent topic distribution related with paper, author, and conference. In this paper, two metadata types are changed. First, conference is replaced with journal. Also, a bag-of-keyphrases are used instead of a bag-of-words to represent documents in a more precise manner.
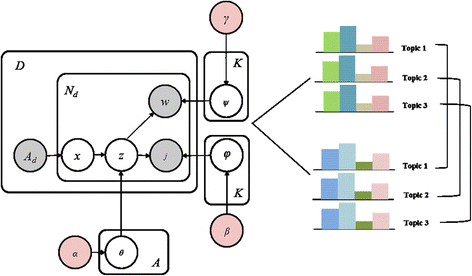
Figure 3 illustrates the ACT model, and Table 4 provides a description of the parameters used. Model estimation is conducted by setting parameters, and for estimation of the model parameter, the Gibbs sampling method is employed. Gibbs sampling takes samples from a probability distribution by using the Markov Chain Monte Carlo sampling method. Three parameters for estimating the model are as follows: 1) θ is the topic probability for a given author (author*topic matrix), 2) φ is the journal probability for a given topic (topic*journal matrix), and 3) ψ is the word probability for a given topic (topic*word matrix). According to the independence assumption, joint distribution of topic, author, journal, and word stand on the basis Ad, meaning the total number of authors in paper d. In our experiments, we set the hyper-parameters α, β, and γ, which are parameters of a prior with α = 50/T, β = 0.01, and γ = 0.01, respectively. In addition, we fix the number of topics K to 20, the number of top keyphrases to 30, and the number of iterations to 1,000. With these settings, we selected 15 out of 20 topics for analysis.
|
| ||||||||||||||||||||||||||||||||
Evaluation
To examine the consistency of our results, we repeated each run 10 times with a topic number of 20. After that, we calculated the similarity between topics. For statistical analysis, we computed Pearson correlation coefficients between any two topics and averaged them out. Table 5 shows the average of correlation coefficients per execution. In all runs, Pearson correlation coefficients between topics were weakly, positively correlated. Also, the range of correlation was not wide (0.13 to 0.18). It implies that there was no difference in similarity between topics regardless of different runs. This result can verify consistency and reliability of our topic clusters.
| ||||||||||||||||||||||||
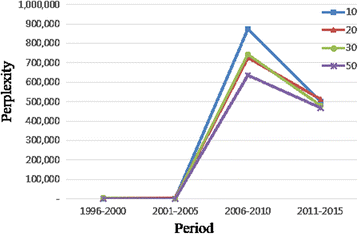
In addition, to evaluate the topic model results, we used perplexity which is a well-known measurement in information theory for testing goodness of a model. In our case, we make a test set by collecting bioinformatics journals published in 2016. The sample size is 1,000 papers. In the training set, we divided 20 years into four periods and calculated the perplexity by setting the number of topics as 10, 20, 30, and 50 respectively. The results are presented in Table 6 and Fig. 4. As shown in Table 6 and also confirmed in Fig. 4, there is not much difference in performance in regards to the number of topics by perplexity. However, there is a clear difference among periods by perplexity. In particular, the third period has the highest perplexity value, which implies that it is the most difficult period as to predicting the topic trend in 2016 in the bioinformatics field.
| ||||||||||||||||||||||||||||||||||||||||||
|
Together with this result, we analyzed the results of the ACT model.
Results
We analyzed leading authors and journals in relation to topics over time. In the following section, we provide the detailed explanations of the trend per period.
Topic analysis per period
The results of our time series topic analysis show that topics seem to be more distinct and subdivided closer to present. In addition, new topics have emerged in recent years, and they do not make a new cluster, which means the exclusive topics become apparent. The results also show that research fields such as molecular biology, genomics, genetics, and proteomics play a supplementary role in biology, but also become diversified into a unique field.
First period analysis
In the first period (1996–2000), five dominant topic clusters were identified (Additional file 1: Appendix 1). Those five topics were mainly associated with proteins and peptides. Phrases such as "molecular biology" and "chemical compound" were widespread, and thermodynamics- and kinematics-related topics appeared. These topics were composed of jargon in their specific fields. The mathematical biology field was represented by topical phrases such as "database," "cluster analysis," "model," "theoretical," and "software."
Topics 0, 2, and 3 were about molecular biology, derived from biochemistry and composed of hydrogen bonding–related chemical compounds such as enzymes or lipids. Topics 4, 5, 6, and 7 were related to proteins, peptides, and protein structure. Topics 9 and 14 included words such as "probability" and "statistics," which are related to mathematical biology. Topics 13, 17, 18, and 19 covered mutagenesis, disease, and syndromes, which are all related to genetic diseases; mutagenesis consists of gene mutation, and syndromes are caused by genetic disorder. Topic 19 included the word "genetic," a parent category of previously mentioned words. Topics 15 and 16 consisted of kinetics.
Protein-related topics were dominant, and authors involved in peptide and protein structure were prevalent in the first period. Authors who were in topic 5, such as A. R. Fersht, J. M. Thornton, C. M. Dobson, L. Serrano, and M. Karplus, had a high probabilistic distribution value, which means they were leading researchers in this area. Their research interest was mainly in protein structure, and they published in the Journal of Molecular Biology. This journal appeared in almost all of the topics related to protein and dealt with structure and function of macromolecules, complexes, and protein folding.
Second period analysis
There were four topic clusters and one exclusive topic in the second period (Additional file 1: Appendix 2). In the second period (2001–2005), studies about genetics and genomics were actively conducted, and protein-related topics were diversified into subfields such as proteomics. In addition, mathematical biology and computational biology–related topics were maintained in this period.
Topics 1, 2, 5, 7, and 11 included DNA mechanisms, molecular structure, genetics, genomics, and diseases caused by DNA or a genome such as Down syndrome, DNA transposable elements, and ribonucleases. Topics 0, 3, 14, and 16 were mainly about proteomics, specifically focusing on protein structure. Topics 12, 18, and 19 addressed biotechnology, molecular modeling, and structure. Topics 8 and 9 focused on mathematical biology and computational biology. Topic 4 exclusively contained enzymology-related phrases such as "enzyme activators" and "oxygen." Enzymology-related topics were less common compared with the first period.
The second period mainly focused on gene-related topics. Topic 5 had the highest probabilistic distribution among top-ranked authors such as Gregory A. Petsko, L. Aravind, Eugene V. Koonin, Mark Gerstein, and Laurence D. Hurst, who were interested in genomics and biomedical engineering. Those authors published papers in Genome Biology, which covered subject matters related to genomics and post-genomics. Similar to the first period, protein-related research was a major topic in the second period. Top-ranked authors in this topic included Ruedi Aebersold, Peter Roepstorff, Pier Giorgio Righetti, Jean-Charles Sanchez, and Peter R. Jungblut. These authors were pioneers of proteomics, and their papers were published in the Journal of Molecular Biology and Proteomics.
Third period analysis
In the third period (2006–2010), topics were divided into three clusters: genomics, proteomics, and other (Additional file 1: Appendix 3). Different from the first two periods, four exclusive topics existed and seemed to be distinct from topics in the other three periods. For instance, studies about genomics or proteomics were more diversified than in the earlier periods. Exclusive topics that were not included in two large fields emerged, indicating that bioinformatics research was conducted in various fields related to bioinformatics.
Topics 3, 7, 10, 11, 13, and 16 consisted of proteomics, protein evolution, and protein structure. Proteomics-related topics were subdivided. The representative journals in the area were Proteomics, the 'Journal of Proteome Research, and the Journal of Proteomics. Topics 5, 6, 12, 14, and 19 were gene-related topics such as gene expression, gene transcription, and genomics. Gene-related studies became prevalent in the second period. The distinct topics that appeared in the third period were topics 0, 15, 17, and 18. Topic 0, molecular biology, especially focused on hydrogen bonding. In the first and second periods, topic 15 included various topics related to theoretical biology. Topic 17 addressed hepatitis, the infection in liver cells and tissues. Different from previous periods, in the third period topics were associated with specific diseases. Topic 18 included peptide-associated phrases, and, unlike prior periods, concrete themes like specific chemical compounds and protein appeared.
Overall, protein-related topics were most common in the third period. The third period also has more sub-divided and distinct topics than previous periods did. In this period, general topics such as proteomics appeared, as did specific topics such as protein evolution, protein analytics, and protein ubiquitin. Among these areas, the topic with the highest distribution was protein analytics, and it was sub-categorized in proteomics. Top-ranked authors in this period included Matthias Mann, Ruedi Aebersold, Richard D. Smith, Albert J. R. Heck, and Visith Thongboonkerd. They were experts in protein analytics and commonly used mass spectrometry for their analyses. They actively published in the Journal of Proteome Research and Proteomics. These two journals were top-rated in protein-related topics. The Journal of Proteome Research was computer technology–oriented and focused on protein-analysis research. The journal with the highest probabilistic distribution in all topic areas was the EMBO Journal. This journal focused on molecular biology and also covered proteomics.
Fourth period analysis
The fourth period (2011–2015) showed three major topic clusters and two exclusive topics (Additional file 1: Appendix 4). Similar to the third period, the topics related with genomics and proteomics were further divided into subfields and represented concrete topical characteristics. Compared with the third period’s results, theoretical biology–related topics formed one cluster. The compositions of the cluster were one big topic (systems biology) and four sub-divided topics.
Topics 1 and 16 were theoretical biology–related, and topics 6 and 10 were about systems biology. They could be clustered as a broader category of system biology. The representative journals in this cluster were PLOS Computational Biology, Journal of Theoretical Biology, and Journal of Computational Neuroscience, which were focused on systems biology. Topics 0, 11, 12, 18, and 19 were about genetics and genomics. Topics 4, 9, 13, and 17 represented proteomics. Topics 8 and 15 were exckusive, each of which was related to molecular biology and cell biology. Topic 8 included phrases like "hydrogen bonding" and "GTP-binding proteins," and topic 15 contained phrases like "enteroendocrine cells" and "COS cells." The top journals in these areas were Biochemistry, the Journal of Molecular Biology, and the Journal of Molecular Modeling.
In the fourth period, the major topics were systems biology, genomics, and proteomics. Topics that were not in the main stream of bioinformatics were found in this period, and topics about theoretical biology and systems biology become a distinct cluster. This means that these areas were growing in the bioinformatics area. The representative researchers in this area were Martin A. Nowak, Yoh Iwasa, Mike Steel, Ulf Dieckmann, and Liam Paninski. They were mostly involved in mathematics and theoretical biology. The journal which had the highest probabilistic distribution was the Journal of Theoretical Biology. This journal focused on research that combines biology and topics such as statistical analysis, mathematical definition, comparative research, experiments, and computer simulation. The second raked journal was the Journal of Bioinformatics, which mainly accepted research about genome bioinformatics and computational biology.
Discussion
In this section, we analyze the results from three different perspectives: topical keyphrase, journal, and author. In addition, to further identify which researchers and journals focus on which topic over time, the results of the ACT model (top-ranked keyphrases, authors, and journals) are examined in an integrated perspective.
Time series analysis
One interesting observation is that keyphrases related with genes or genetic processes such as "gene expression," "down-regulation," and "up-regulation" were not ranked high in the first period. However, they emerged as top keyphrases in later periods. In particular, "proteome," "reproducibility of results," "proteomics," and "genotype" did not appear in the first period but emerged gradually after that. In addition, the term "algorithms" appeared most frequently in the fourth period. In author perspective, across the four periods, the number of unique authors was 1,396. Top ranked author Richard Robinson appeared in five topics. Seven authors, including Liza Gross, appeared four times, 45 authors appeared three times, 137 authors appeared twice, and the remaining 1,184 authors were shown only in one topic. There was no author who appeared in all four periods. Thirty-nine authors appeared across three periods, 125 authors appeared in two periods, and 1,210 authors appeared only in a period. In journal centered view, only 21 out of 46 journals appeared in the first period. In the second and third period, 34 and 46 journals were presented respectively. Forty-five journals appeared in the last period; one the journals, Briefings in Functional Genomics & Proteomics, did not appear in the last period.
These results imply that the bioinformatics field is diversified and new topical disciplines have recently emerged. For instance, proteomics-related topics started to appear in the second period, became segmented into detailed research fields and later evolved in the third and fourth periods. In addition, while conceptual biology–related topics existed in the first period, they became clearly progressed in the fourth period. Conversely, the topics about kinetics appeared in the first period but then faded out.
Integrated view of graph pattern analysis
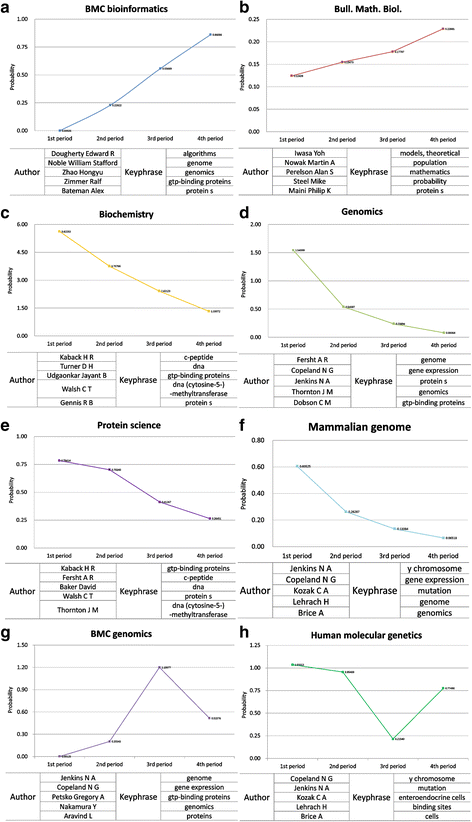
For further integrated analysis, we examined top journals with their topical probability in all four periods. We also checked the authors and topical keyphrases which were topically matched with the journals. We identified that there were four different patterns in a journal’s topical distribution such as rising, falling, concave, and convex patterns. In Fig. 5, we only presented graphs which drastically changed in terms of the probability value of topics between periods. Additionally in each graph, we presented the top five ranked authors and keyphrases which had a high probability value across four periods.
|
We explained four outstanding cases in each pattern and made a list of journals which are shown in all four periods (Table 7). First for the rising pattern, the journal BMC Bioinformatics had 0.86060 gaps between maximum and minimum probability. It was the highest gap by probability in all rising patterns. The average impact factor of this journal provided by journal citation report (JCR) was 3.0806 in 2015. In this context, BMC Bioinformatics could be regarded as the most promising journal in the bioinformatics field. The journal has grown steadily through 20 years. The authors belonging to the similar topical scope of BMC Bioinformatics were presented in graph (a). The top ranked authors shared common characteristics: the authors were majored in computer science or statistics and then applied the technique to biomedical or biology later on, and their common research interests were bioinformatics or biostatistics. As shown in graph (a), the topical keyphrases related with the journal were not focused only on biology research fields. The word "algorithms" represents the informatics field, while "genome," "genomics," and "gtp-binding proteins" relate to the genomics fields, and the word "protein s" represents protein- or gene-related scopes. The scope of the journal is in computational and statistical methods for the modeling and analysis of all kinds of biological data, as well as other areas of computational biology. The results indicated that the publication trend of BMC Bioinformatics changed to genetics or genomics converged with informatics.
In the case of the falling pattern of the journal Biochemistry, as shown in graph (c), the journal had 4.29181 gap between the maximum and minimum value. The average impact factor of this journal in four periods was 3.75322. The journal had a somewhat high probability value, but the impact factor in each period decreased gradually (e.g., first period: 4.4785 to fourth period: 3.1768). This decreasing pattern implied that in bioinformatics field, the journal dealt mainly with biochemistry, biophysical chemistry, and molecular biology, but it had not converged with informatics. That means the biochemistry field did not have much interaction with informatics fields.
The top ranked authors shown in graph (c) were celebrated scholars in the biochemistry field. Commonly, they all researched only the biochemistry field, and we could not find that they had a connection with the informatics fields. In the same context, top ranked keyphrases of the journal were mostly related to biochemistry research fields. The informatics-related keyphrase did not appear in the top word list.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Not only the rising and falling patterns, but the convex and concave patterns of journals exist. In graph (g), BMC Genomics was the journal showing the convex pattern. BMC Genomics dealt with genome-scale analysis, functional genomics, and proteomics. The average impact factor from 2000 to 2015 was 4.0464. The topical probability of the journal rose from the first period (0.00125) to the third period (1.19977), but since then the probability dropped to 0.51576. The gap between maximum and minimum value was 1.19852. The journal changed the status to open-access in 2000, and maybe it caused a slight rise from first period to second period. For the drastic increase from second period to third period, the number of total citations increased approximately two times according to the JCR. The increasing and decreasing number in total citations may cause the wave in graph (g). The top five ranked authors’ research interests had various characteristics. N. A. Jenkins and N.G. Copeland were a couple, with Jenkins interested in molecular and cellular biology, while the husband Copeland was in the biochemistry field. They collaborated a lot and both were celebrated researchers in the field. The other researchers also had different research areas, including treatment or preventive therapy (Gregory A. Petsko), genomic medicine (Y. Nakamura), and genome analysis (L. Aravind). Through all four periods, the journal mainly published articles which dealt with the genomic related keyphrases. The authors and keyphrases located in this journal were not quite related to the informatics fields.
Last, the concave pattern was shown in graph (h), and Human Molecular Genetics was the representative journal. The journal had steadily decreasing impact factor flow in 20 years (e.g., first period: 9.05475 to fourth period: 6.8766). The gap between maximum and minimum probability was 0.81973. In graph (h), the point of drastic decrease can be readily observed. Top ranked authors related with the journal focused on genetics. N. G. Copeland and N. A. Jenkins appeared again. The rest of the authors did not overlap in research areas. The journal was interested in broad genetics-related topical keyphrases but not related with informatics fields.
Through the pattern analysis integrated with journal, author, and keyphrase, we identified that the field of bioinformatics was a converging area, and certain journals clearly showed rising and falling patterns. Different from period analysis, the integrated view of analysis showed journals’ topical trends over time along with top journals and researchers.
Conclusions
Bioinformatics mainly tackles biological problems at the molecular level using applied mathematics, information science, statistics, computer science, chemistry, and biochemistry. This characteristic of bioinformatics has driven the field to become interdisciplinary, combining approaches from various fields to make use of a large amount of data.
In this study, we investigated the bioinformatics field using the ACT model to conduct comprehensive topic analyses of keyphrases, authors, and journals. To this end, we collected 46 journals belonging to the bioinformatics field by searching journal name in PubMed, yielding 170,099 papers. To analyze topic evolution over time, we divided the collected datasets into four, five-year periods: 1996–2000, 2001–2005, 2006–2010, and 2011–2015. In a time series topic analysis, we examined topic clusters within periods. In more recent periods, distinct characteristics of the field emerged and more new topics were presented independently. In addition, we analyzed trends in keyphrases, authors, and journals. Our keyphrase analysis similarly indicated the emergence of greater interdisciplinary research over time. In our author analysis, we observed the pattern of authors who appear in top rank. In our journal analysis, we analyzed the common topic area of top journals and identified major focus areas of those top journals, including computational biology, theoretical biology, and mathematical biology. Also, we examined topic distribution over journals with top ranked authors and keyphrases. In this analysis, journals were identified as four patterns over time such as rising, falling, convex, and concave patterns.
The results of these analyses imply that the bioinformatics field is highly interdisciplinary, consisting of active convergence studies. In addition, we observed that the characteristics of the bioinformatics field become more distinct and more specific, and the supporting role of peripheral fields of bioinformatics, such as conceptual, mathematical, and systems biology, gradually increases over time, though the core fields of proteomics, genomics, and genetics are still the major topics. This is consistently confirmed by analysis of topic distribution of journals over time as well as integrated analysis of top ranked keyphrases, authors, and journals.
In the future, we plan to apply the same approach to other domains, such as information science. We also plan to use other metadata such as MeSH terms and bio-entities to compare with keyphrases. In addition, we plan to explore how to infer authors’ interests by time series analysis and identify representative authors and the journals that are the best suited for a paper on a particular subject.
Declarations
Acknowledgements
Not applicable.
Funding
This work was supported by the Bio-Synergy Research Project (NRF-2013M3A9C4078138) of the Ministry of Science, ICT and Future Planning through the National Research Foundation. This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. NRF-2014R1A2A2A01004454). The publication charges for this article was funded by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP).
Availability of data and materials
Not applicable.
Authors' contributions
GEH conceived and designed the study. GEH and KYK conducted the analysis and drafted the manuscript. MS designed and coordinated the study. All authors contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
About this supplement
This article has been published as part of BMC Bioinformatics Volume 18 Supplement 7, 2017: Proceedings of the Tenth International Workshop on Data and Text Mining in Biomedical Informatics. The full contents of the supplement are available online at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-18-supplement-7.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
- Additional File 1: Appendix 1. ACT model results in first period. Appendix 2. ACT model results in second period. Appendix 3. ACT model results in third period. Appendix 4. ACT model results in fourth period. (DOCX 48 kb)
References
- ↑ Dong, D.; Chen, M.-L. (2015). "Publication trends and co-citation mapping of translation studies between 2000 and 2015". Scientometrics 105 (2): 1111–1128. doi:10.1007/s11192-015-1769-1.
- ↑ 2.0 2.1 Chen, H.; Wan, Y.; Jiang, S.; Cheng, Y. (2014). "Alzheimer’s disease research in the future: bibliometric analysis of cholinesterase inhibitors from 1993 to 2012". Scientometrics 98 (3): 1865–1877. doi:10.1007/s11192-013-1132-3.
- ↑ 3.0 3.1 Soteriades, E.S.; Falagas, M.E. (2006). "Alzheimer’s disease research in the future: Bibliometric analysis of cholinesterase inhibitors from 1993 to 2012". BMC Public Health 6: 301. doi:10.1186/1471-2458-6-301. PMC PMC1766930. PMID 17173665. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1766930.
- ↑ 4.0 4.1 Ugolini, D.; Puntoni, R.; Perera, F.P. et al. (2007). "A bibliometric analysis of scientific production in cancer molecular epidemiology". Carcinogenesis 28: 8. doi:10.1093/carcin/bgm129. PMID 17548902.
- ↑ Wang, L.; Chen, X.; Bao, A. et al. (2015). "A bibliometric analysis of research on Central Asia during 1990–2014". Scientometrics 105 (2): 1223–1237. doi:10.1007/s11192-015-1727-y.
- ↑ 6.0 6.1 Bornmann, L.; Mutz, R. (2015). "Growth rates of modern science: A bibliometric analysis based on the number of publications and cited references". Journal of the Association for Information Science and Technology 66 (11): 2215–2222. doi:10.1002/asi.23329.
- ↑ 7.0 7.1 Geaney, F.; Scutaru, C.; Kelly, C. et al. (2015). "Type 2 Diabetes Research Yield, 1951-2012: Bibliometrics Analysis and Density-Equalizing Mapping". PLoS One 10: 7. doi:10.1371/journal.pone.0133009. PMC PMC4514795. PMID 26208117. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4514795.
- ↑ 8.0 8.1 Macías-Chapula, C.A.; Mijangos-Nolasco, A. (2002). "Bibliometric analysis of AIDS literature in Central Africa". Scientometrics 54 (2): 309–317. doi:10.1023/A:1016074230843.
- ↑ 9.0 9.1 Seglen, P.O.; Aksnes, D.W. (2000). "Scientific Productivity and Group Size: A Bibliometric Analysis of Norwegian Microbiological Research". Scientometrics 49 (1): 125–143. doi:10.1023/A:1005665309719.
- ↑ 10.0 10.1 Jeong, D.-H.; Song, M. (2014). "Time gap analysis by the topic model-based temporal technique". Journal of Infometrics 8 (3): 776–790. doi:10.1016/j.joi.2014.07.005.
- ↑ 11.0 11.1 11.2 Song, M.; Kim, S.Y. (2013). "Detecting the knowledge structure of bioinformatics by mining full-text collections". Scientometrics 96 (1): 183–201. doi:10.1007/s11192-012-0900-9.
- ↑ 12.0 12.1 Song, M.; Kim, S.Y.; Zhang, G. et al. (2014). "Productivity and influence in bioinformatics: A bibliometric analysis using PubMed central". Journal of the Association for Information Science and Technology 65 (2): 352–371. doi:10.1002/asi.22970.
- ↑ 13.0 13.1 Yan, E. (2015). "Research dynamics, impact, and dissemination: A topic-level analysis". Journal of the Association for Information Science and Technology 66 (1): 2357–2372. doi:10.1002/asi.23324.
- ↑ 14.0 14.1 Steyvers, M.; Smyth, P.; Rosen-Zvi, M.; Griffiths, T. (2004). "Probabilistic author-topic models for information discovery". Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2004: 306–315. doi:10.1145/1014052.1014087.
- ↑ 15.0 15.1 Li, D.; Okamoto, J.; Liu, H.; Leischow, S. (2015). "A bibliometric analysis on tobacco regulation investigators". BioData Mining 8: 11. doi:10.1186/s13040-015-0043-7. PMC PMC4432889. PMID 25984237. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4432889.
- ↑ 16.0 16.1 Rosen-Zvi, M.; Griffiths, T., Steyvers, M.; Smyth, P. (2004). "The author-topic model for authors and documents". Proceedings of the 20th Conference in Uncertainty in Artificial Intelligence 2004: 487–494. ISBN 0974903906.
- ↑ 17.0 17.1 17.2 Tang, J.; Zhang, J.; Yao, L. et al. (2008). "ArnetMiner: Extraction and mining of academic social networks". Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2008: 990-998. doi:10.1145/1401890.1402008.
- ↑ Ramos, J.M.; Padilla, S.; Masiá, M.; Gutiérrez, F. (2008). "A bibliometric analysis of tuberculosis research indexed in PubMed, 1997-2006". International Journal of Tuberculosis and Lung Disease 2008: 1461–8. PMID 19017458.
- ↑ Claude, R.; Charles-Daniel, A.; Jean, A.; Jean-Francois, G. (2004). "Bibliometric overview of the utilization of artificial neural networks in medicine and biology". Scientometrics 59 (1): 117–130. doi:10.1023/B:SCIE.0000013302.59845.34.
- ↑ Patra, S.K.; Mishra, S. (2006). "Bibliometric study of bioinformatics literature". Scientometrics 67 (3): 477–489. doi:10.1556/Scient.67.2006.3.9.
- ↑ Song, M.; Heo, G.E.; Kim, S.Y. (2014). "Analyzing topic evolution in bioinformatics: Investigation of dynamics of the field with conference data in DBLP". Scientometrics 101 (1): 397–428. doi:10.1007/s11192-014-1246-2.
- ↑ Lee, D.; Kim, W.C.; Charidimou, A.; Song, M. (2015). "A Bird's-Eye View of Alzheimer's Disease Research: Reflecting Different Perspectives of Indexers, Authors, or Citers in Mapping the Field". Journal of Alzheimer's Disease 45 (4): 1207-22. doi:10.3233/JAD-142688. PMID 25697702.
- ↑ Ding, Y.; Song, M.; Han, J. et al. (2013). "Entitymetrics: Measuring the impact of entities". PLoS One 8 (8): e71416. doi:10.1371/journal.pone.0071416. PMC PMC3756961. PMID 24009660. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3756961.
- ↑ Hofmann, T. (1999). "Probabilistic latent semantic indexing". Proceedings of the 22nd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval 1999: 50–57. doi:10.1145/312624.312649.
- ↑ 25.0 25.1 Blei, D.M.; Ng, A.Y.; Jordan, M.I. (2003). "Latent dirichlet allocation". Journal of Machine Learning Research 3 (1): 993–1022. http://jmlr.csail.mit.edu/papers/v3/blei03a.html.
- ↑ Tang, J.; Zhang, J.; Jin, R. et al. (2011). "Topic level expertise search over heterogeneous networks". Machine Learning 82 (2): 211–237. doi:10.1007/s10994-010-5212-9.
- ↑ Kim, H.J.; An, J.; Jeong, Y.K.; Song, M. (2016). "Exploring the Leading Authors and Journals in Major Topics by Citation Sentences and Topic Modeling". Proceedings of the Joint Workshop on Bibliometric-enhanced Information Retrieval and Natural Language Processing for Digital Libraries (BIRNDL) co-located with the Joint Conference on Digital Libraries 2016 2016: 42–50. http://dblp.uni-trier.de/db/conf/jcdl/birndl2016.html.
- ↑ Medelyan, O. (2009). "Human-competitive automatic topic indexing (Thesis)". The University of Waikato. http://hdl.handle.net/10289/3513.
- ↑ Witten, I.H.; Paynter, G.W.; Frank, E. et al. (1999). "KEA: Practical automatic keyphrase extraction". Proceedings of the Fourth ACM Conference on Digital Libraries 1999: 254–255. doi:10.1145/313238.313437.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Grammar and word used were updated to make the text easier to read.