Difference between revisions of "Main Page/Featured article of the week/2022"
Shawndouglas (talk | contribs) (Added last week's article of the week) |
Shawndouglas (talk | contribs) |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{ombox | {{ombox | ||
| type = notice | | type = notice | ||
| text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - [[Main Page/Featured article of the week/2017|2017]] - [[Main Page/Featured article of the week/2018|2018]] - [[Main Page/Featured article of the week/2019|2019]] - [[Main Page/Featured article of the week/2020|2020]] - [[Main Page/Featured article of the week/2021|2021]] - 2022 | | text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - [[Main Page/Featured article of the week/2017|2017]] - [[Main Page/Featured article of the week/2018|2018]] - [[Main Page/Featured article of the week/2019|2019]] - [[Main Page/Featured article of the week/2020|2020]] - [[Main Page/Featured article of the week/2021|2021]] - 2022 - [[Main Page/Featured article of the week/2023|2023]] - [[Main Page/Featured article of the week/2024|2024]] | ||
}} | }} | ||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
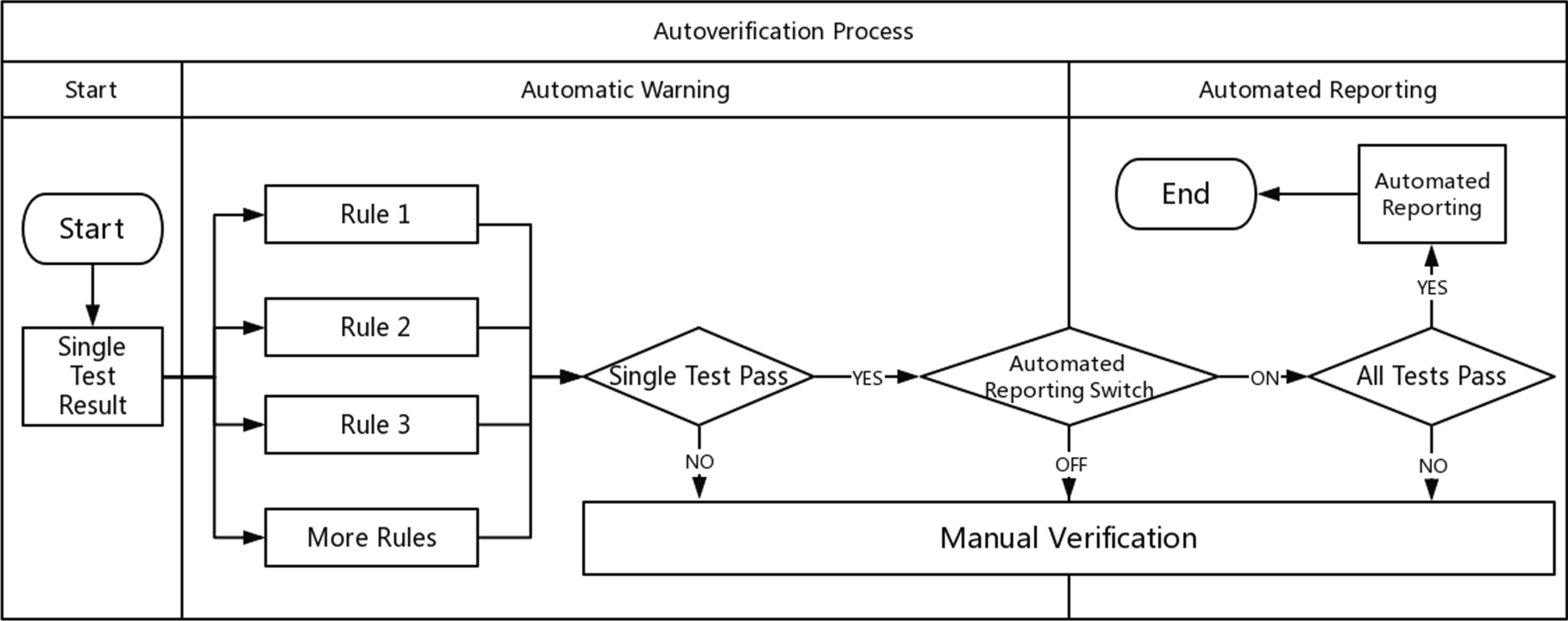
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 25–31:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 19–25:</h2> | ||
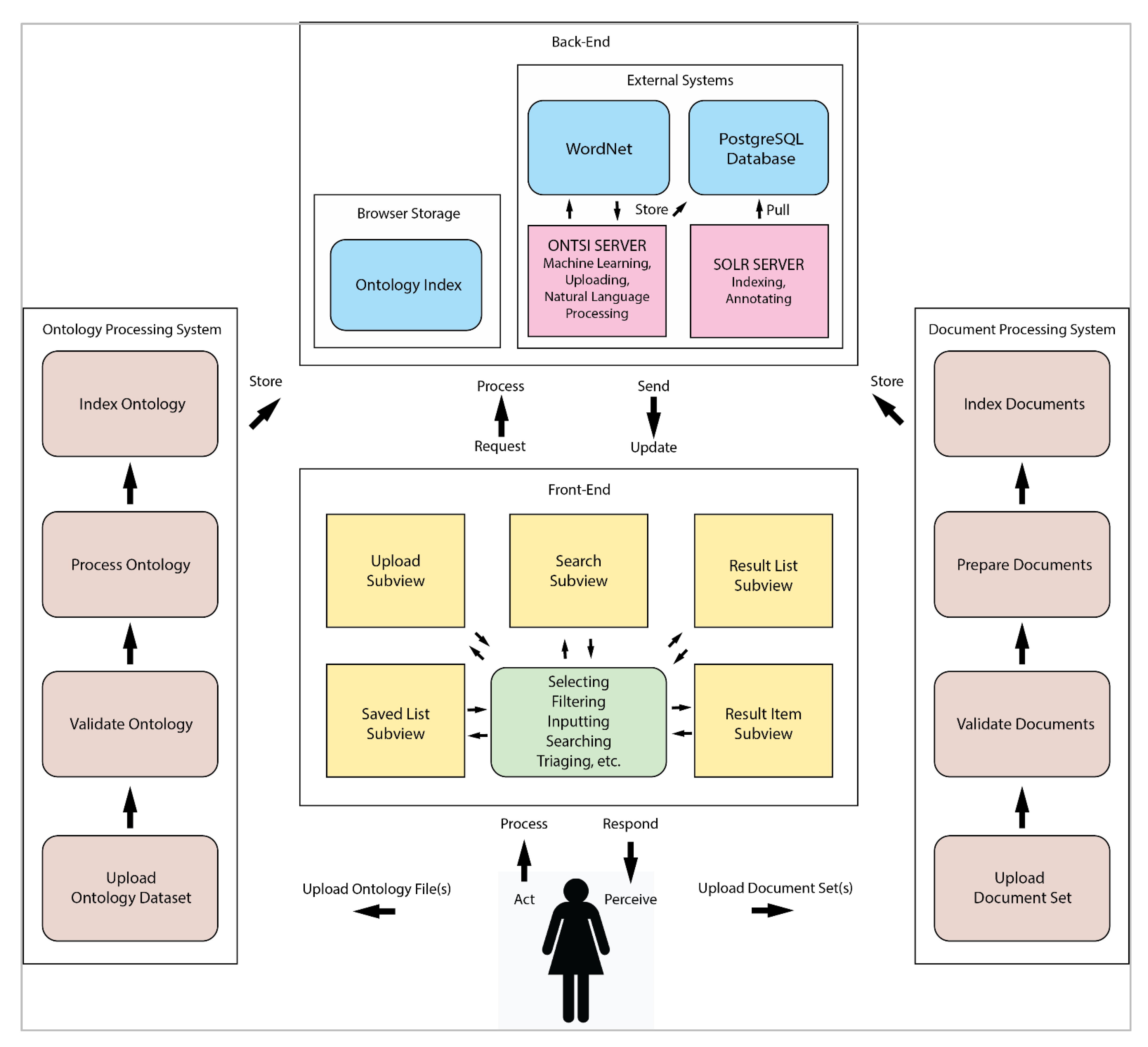
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Cecchetti JMIRMedInfo8 8-10.png|240px]]</div> | |||
'''"[[Journal:Fueling clinical and translational research in Appalachia: Informatics platform approach|Fueling clinical and translational research in Appalachia: Informatics platform approach]]"''' | |||
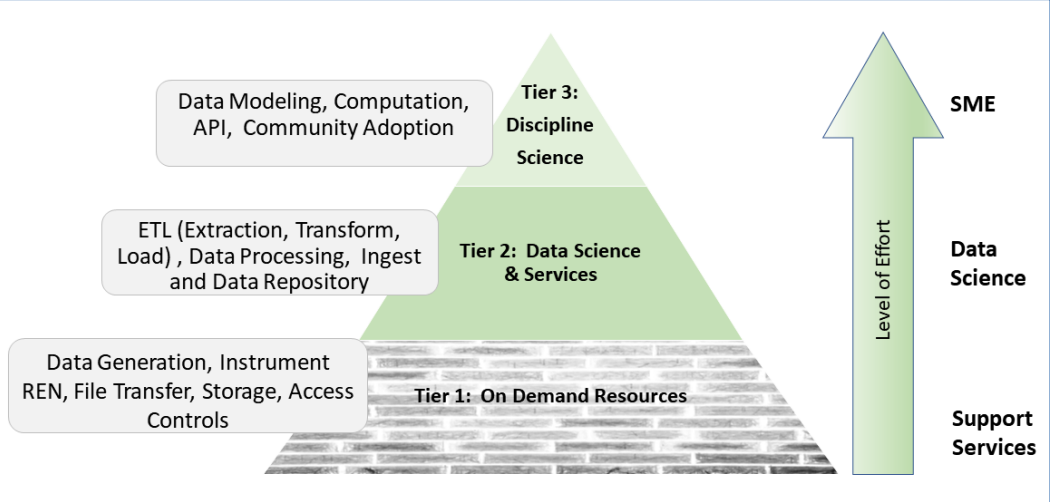
The Appalachian population is distinct, not just culturally and geographically but also in its healthcare needs, facing the most health care disparities in the United States. To meet these unique demands, Appalachian medical centers need an arsenal of analytics and data science tools with the foundation of a centralized [[data warehouse]] to transform healthcare data into actionable clinical interventions. However, this is an especially challenging task given the fragmented state of medical data within Appalachia and the need for integration of other types of data such as environmental, social, and economic with medical data. This paper aims to present the structure and process of the development of an integrated platform at a midlevel Appalachian academic medical center, along with its initial uses ... ('''[[Journal:Fueling clinical and translational research in Appalachia: Informatics platform approach|Full article...]]''')<br /> | |||
|- | |||
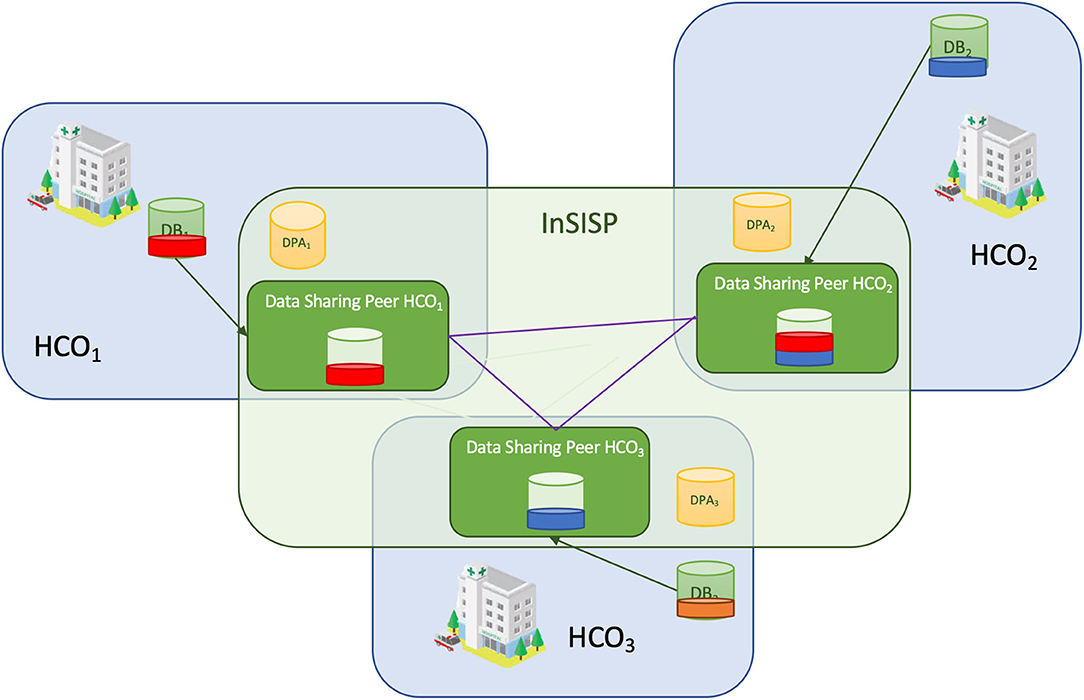
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 12–18:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Sachdeva Information22 13-2.png|240px]]</div> | |||
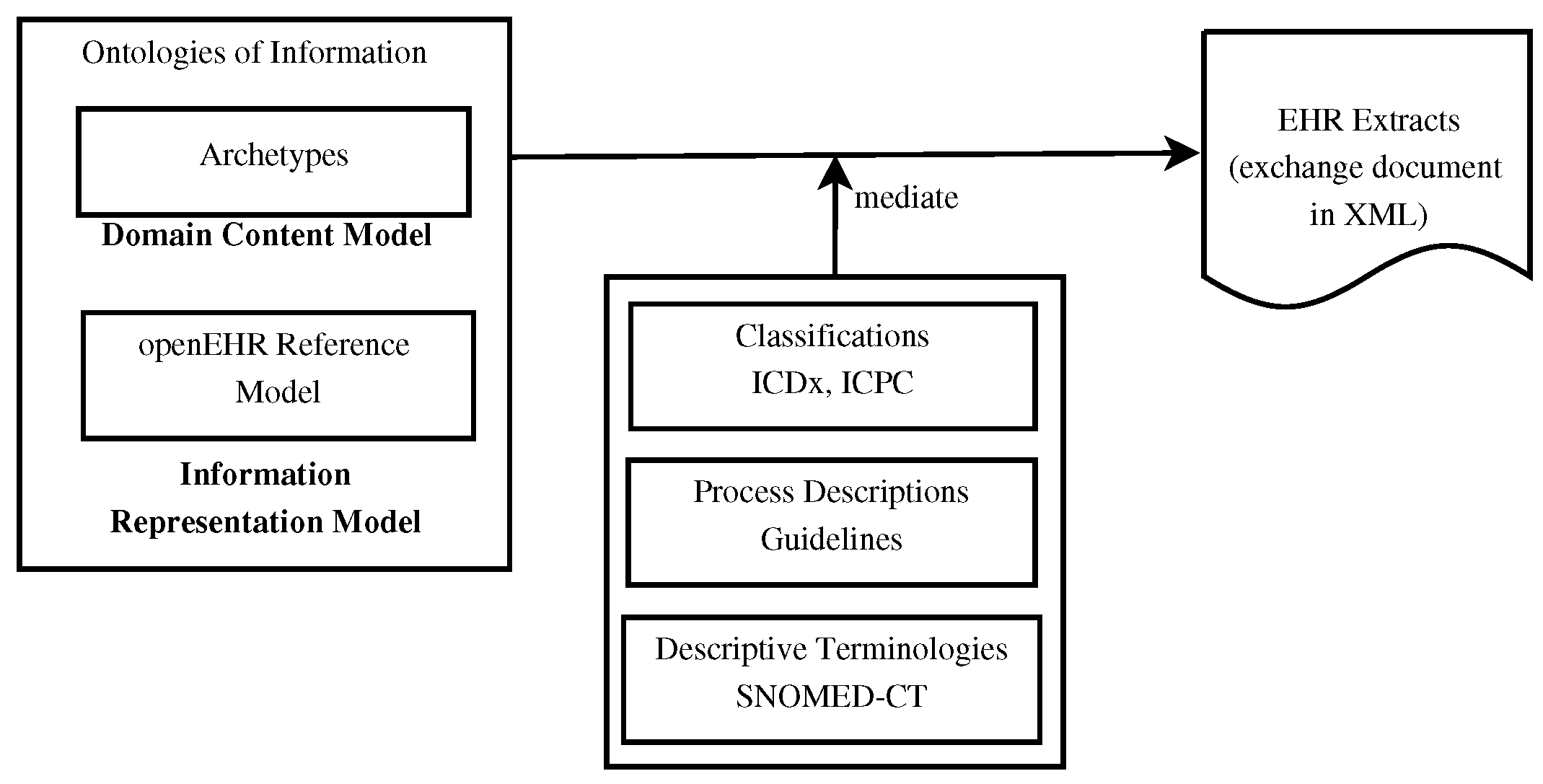
'''"[[Journal:Using knowledge graph structures for semantic interoperability in electronic health records data exchanges|Using knowledge graph structures for semantic interoperability in electronic health records data exchanges]]"''' | |||
[[Data sharing|Information sharing]] across medical institutions is restricted to [[information]] exchange between specific partners. The lifelong [[electronic health record]] (EHR) structure and content require standardization efforts. Existing standards such as [[openEHR]], [[Health Level 7]] (HL7), and ISO/EN 13606 aim to achieve data independence along with semantic interoperability. This study aims to discover knowledge representation to achieve semantic health data exchange. openEHR and ISO/EN 13606 use archetype-based technology for semantic interoperability. The HL7 Clinical Document Architecture is on its way to adopting this through HL7 templates. Archetypes are the basis for knowledge-based systems, as these are means to define clinical knowledge. The paper examines a set of formalisms for the suitability of describing, representing, and reasoning about archetypes. Each of the information exchange technologies—such as XML, Web Ontology Language (OWL), Object Constraint Language (OCL), and Knowledge Interchange Format (KIF)—is evaluated as a part of the knowledge representation experiment. These examine the representation of archetypes as described by Archetype Definition Language (ADL) ... ('''[[Journal:Using knowledge graph structures for semantic interoperability in electronic health records data exchanges|Full article...]]''')<br /> | |||
|- | |||
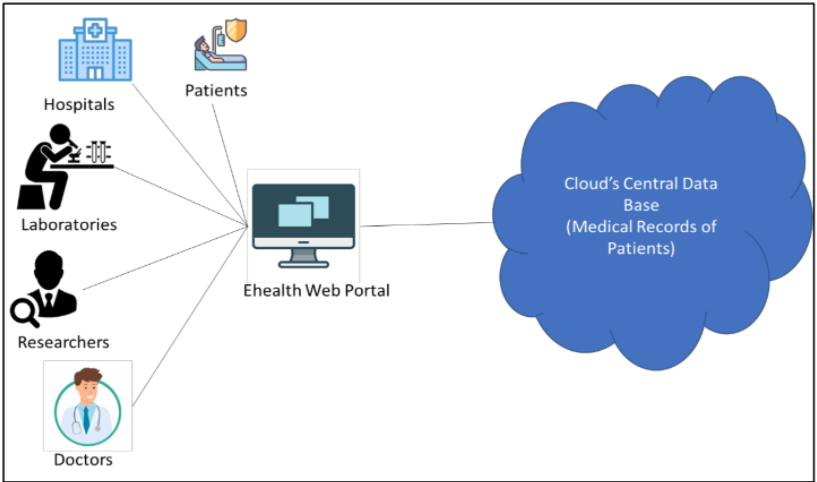
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 05–11:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Mwambe IntJofAdvSciResEng22 8-4.png|240px]]</div> | |||
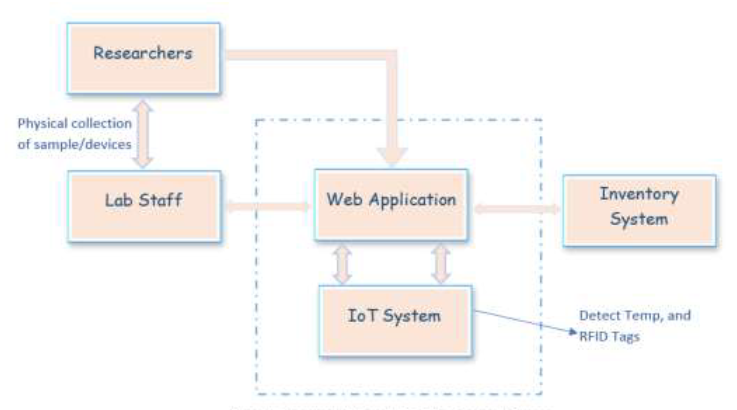
'''"[[Journal:Development of a smart laboratory information management system: A case study of NM-AIST Arusha of Tanzania|Development of a smart laboratory information management system: A case study of NM-AIST Arusha of Tanzania]]"''' | |||
Testing laboratories in higher learning institutions of science, technology, and engineering are used by institutional staff, researchers, and external stakeholders in conducting research experiments, [[Sample (material)|sample]] analysis, and result dissemination. However, there exists a challenge in the management of [[laboratory]] operations and processing of laboratory-based data. Operations carried out in the laboratory at Nelson Mandela African Institution of Science and Technology (NM-AIST), in Arusha, Tanzania—where this case study was carried out—are paper-based. There is no automated way of sample registration and identification, and researchers are prone to making errors when handling sensitive reagents. Users have to physically visit the laboratory to enquire about available equipment or reagents before borrowing or reserving those resources. Additionally, paper-based forms have to be filled out and handed to the laboratory manager for approval ... ('''[[Journal:Development of a smart laboratory information management system: A case study of NM-AIST Arusha of Tanzania|Full article...]]''')<br /> | |||
|- | |||
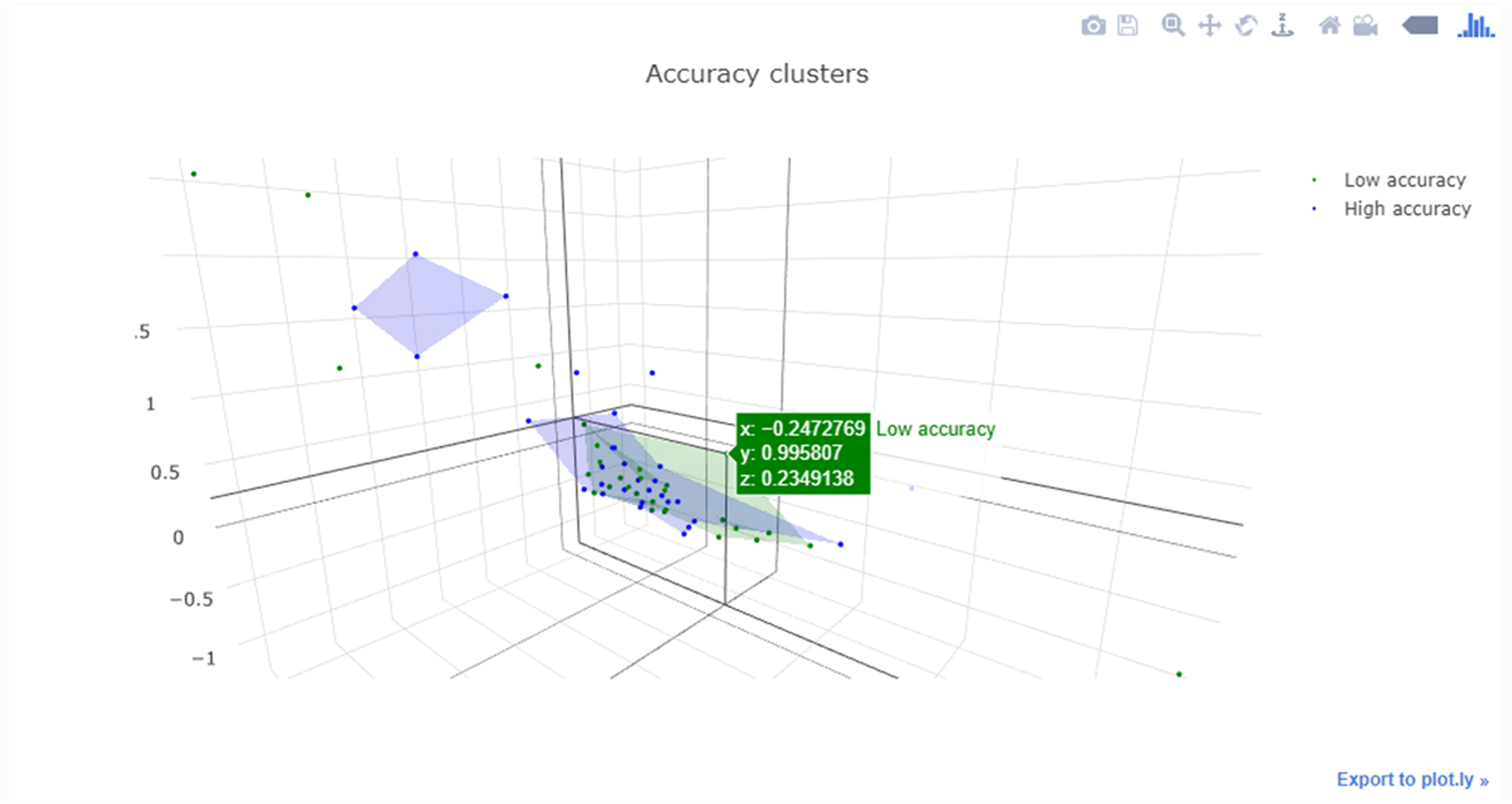
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 28–December 04:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Yogesh FrontPubHlth2022 10.jpg|240px]]</div> | |||
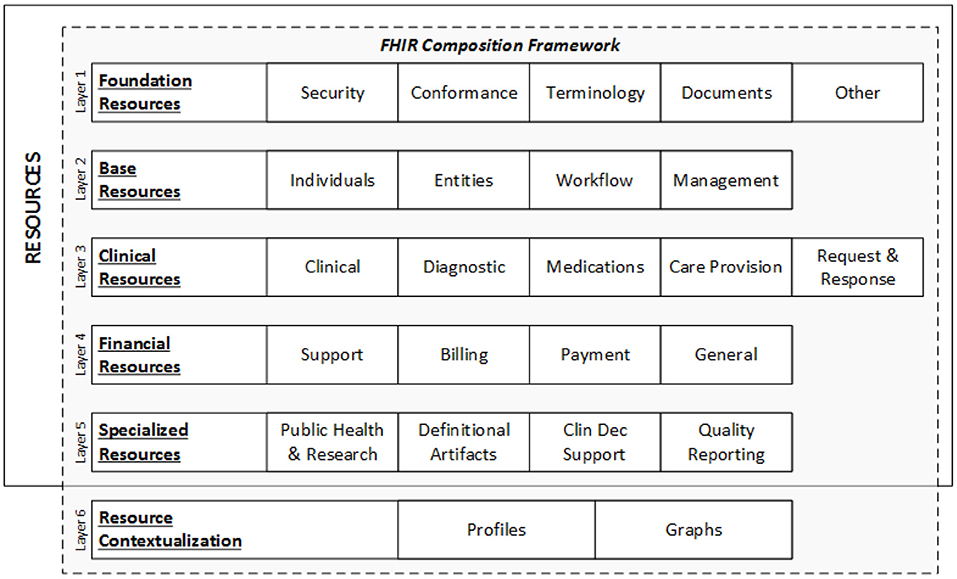
'''"[[Journal:Health informatics: Engaging modern healthcare units: A brief overview|Health informatics: Engaging modern healthcare units: A brief overview]]"''' | |||
With a large amount of unstructured data finding its way into health systems, [[health informatics]] implementations are currently gaining traction, allowing healthcare units to leverage and make meaningful insights for doctors and decision makers using relevant [[information]] to scale operations and predict the future view of treatments via information systems communication. Now, around the world, massive amounts of data are being collected and analyzed for better patient diagnosis and treatment, improving [[public health]] systems and assisting government agencies in designing and implementing public health policies, while also instilling confidence in future generations who want to use better public health systems. This article provides an overview of the [[Health Level 7#Fast Healthcare Interoperability Resources (FHIR)|Health Level 7 FHIR]] architecture ... ('''[[Journal:Health informatics: Engaging modern healthcare units: A brief overview|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 21–27:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Green NIST221-22.png|240px]]</div> | |||
'''"[[Journal:A roadmap for LIMS at NIST Material Measurement Laboratory|A roadmap for LIMS at NIST Material Measurement Laboratory]]"''' | |||
Over the past decade, emerging technology in [[laboratory]] and computational science has changed the landscape for research by accelerating the production, processing, and exchange of data. The NIST Material Measurement Laboratory community recognizes that to keep pace with the transformation of measurement science to a digital paradigm, it is essential to implement [[laboratory information management system]]s (LIMS). Effective introduction of LIMS early in the research life cycle provides direct support for planning and execution of experiments and accelerating research productivity. From this perspective, LIMS are not passive entities with isolated interaction, but rather key resources supporting collaboration, scientific integrity, and transfer of knowledge over time. They serve as a delivery system for organizational contributions to the broader federated data community, supporting both controlled and open access, determined by the sensitivity of the research ... ('''[[Journal:A roadmap for LIMS at NIST Material Measurement Laboratory|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 14–20:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Tomlinson JofMolDiag2022 S1525-1578-22.jpg|240px]]</div> | |||
'''"[[Journal:A model for design and implementation of a laboratory information management system specific to molecular pathology laboratory operations|A model for design and implementation of a laboratory information management system specific to molecular pathology laboratory operations]]"''' | |||
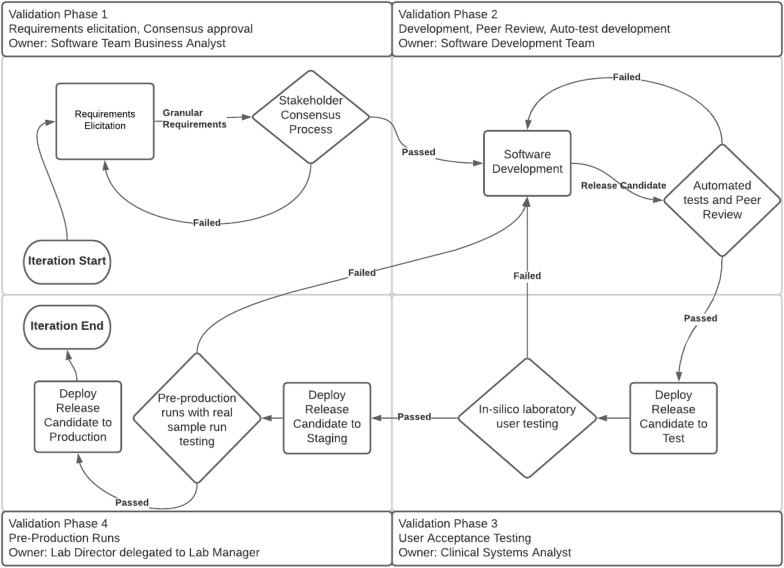
The Molecular Pathology Section of Cleveland Clinic (Cleveland, OH) has undergone enhancement of its testing portfolio and processes. An electronic- and paper-based [[Information management|data management]] system was replaced with a commercially available [[laboratory information management system]] (LIMS) solution, a separate [[bioinformatics]] platform, customized test-interpretation applications, a dedicated [[Accessioning (medical)|accessioning]] service, and a results-releasing solution. The LIMS solution manages complex [[workflow]]s, large-scale data packets, and process automation. However, a customized approach was required for the LIMS since a survey of commercially available off-the-shelf (COTS) software solutions revealed none met the diverse and complex needs of Cleveland Clinic's [[molecular pathology]] service ... ('''[[Journal:A model for design and implementation of a laboratory information management system specific to molecular pathology laboratory operations|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 7–13:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig4 Temprana-Salvado Diagnostics22 12-4.png|240px]]</div> | |||
'''"[[Journal:DigiPatICS: Digital pathology transformation of the Catalan Health Institute network of eight hospitals - Planning, implementation, and preliminary results|DigiPatICS: Digital pathology transformation of the Catalan Health Institute network of eight hospitals - Planning, implementation, and preliminary results]]"''' | |||
Complete [[digital pathology]] transformation for primary [[Histopathology|histopathological]] diagnosis is a challenging yet rewarding endeavor. Its advantages are clear with more efficient [[workflow]]s, but there are many technical and functional difficulties to be faced. The Catalan Health Institute (Institut Català de la Salut or ICS) has started its DigiPatICS project, aiming to deploy digital pathology in an integrative, holistic, and comprehensive way within a network of eight [[hospital]]s, over 168 [[Pathology|pathologists]], and over one million slides each year. We describe the bidding process and the careful planning that was required, followed by swift implementation in stages. The purpose of the DigiPatICS project is to increase patient safety and quality of care, improving diagnosis and the efficiency of processes in the pathological anatomy departments of the ICS through process improvement, digital pathology, and [[artificial intelligence]] (AI) tools ... ('''[[Journal:DigiPatICS: Digital pathology transformation of the Catalan Health Institute network of eight hospitals - Planning, implementation, and preliminary results|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 31–November 6:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Schröder JofBioSem22 13.png|240px]]</div> | |||
'''"[[Journal:Structure-based knowledge acquisition from electronic lab notebooks for research data provenance documentation|Structure-based knowledge acquisition from electronic lab notebooks for research data provenance documentation]]"''' | |||
[[Electronic laboratory notebook]]s (ELNs) are used to document experiments and investigations in the wet [[Laboratory|lab]]. Protocols in ELNs contain a detailed description of the conducted steps, including the necessary [[information]] to understand the procedure and the raised research data, as well as to reproduce the research investigation. The purpose of this study is to investigate whether such ELN protocols can be used to create [[wikipedia:Semantics|semantic]] documentation of the provenance of research data by the use of [[Ontology (information science)|ontologies]] and linked data methodologies. Based on an ELN protocol of a biomedical wet lab experiment, a retrospective [[wikipedia:Provenance#Data provenance|provenance model]] of the raised research data describing the details of the experiment in a machine-interpretable way is manually engineered. Furthermore, an automated approach for knowledge acquisition from ELN protocols is derived from these results. This structure-based approach exploits the structure in the experiment’s description—such as headings, tables, and links—to translate the ELN protocol into a semantic knowledge representation ... ('''[[Journal:Structure-based knowledge acquisition from electronic lab notebooks for research data provenance documentation|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 24–30:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Krupitzer Foods21 10-11.png|240px]]</div> | |||
'''"[[Journal:Food informatics: Review of the current state-of-the-art, revised definition, and classification into the research landscape|Food informatics: Review of the current state-of-the-art, revised definition, and classification into the research landscape]]"''' | |||
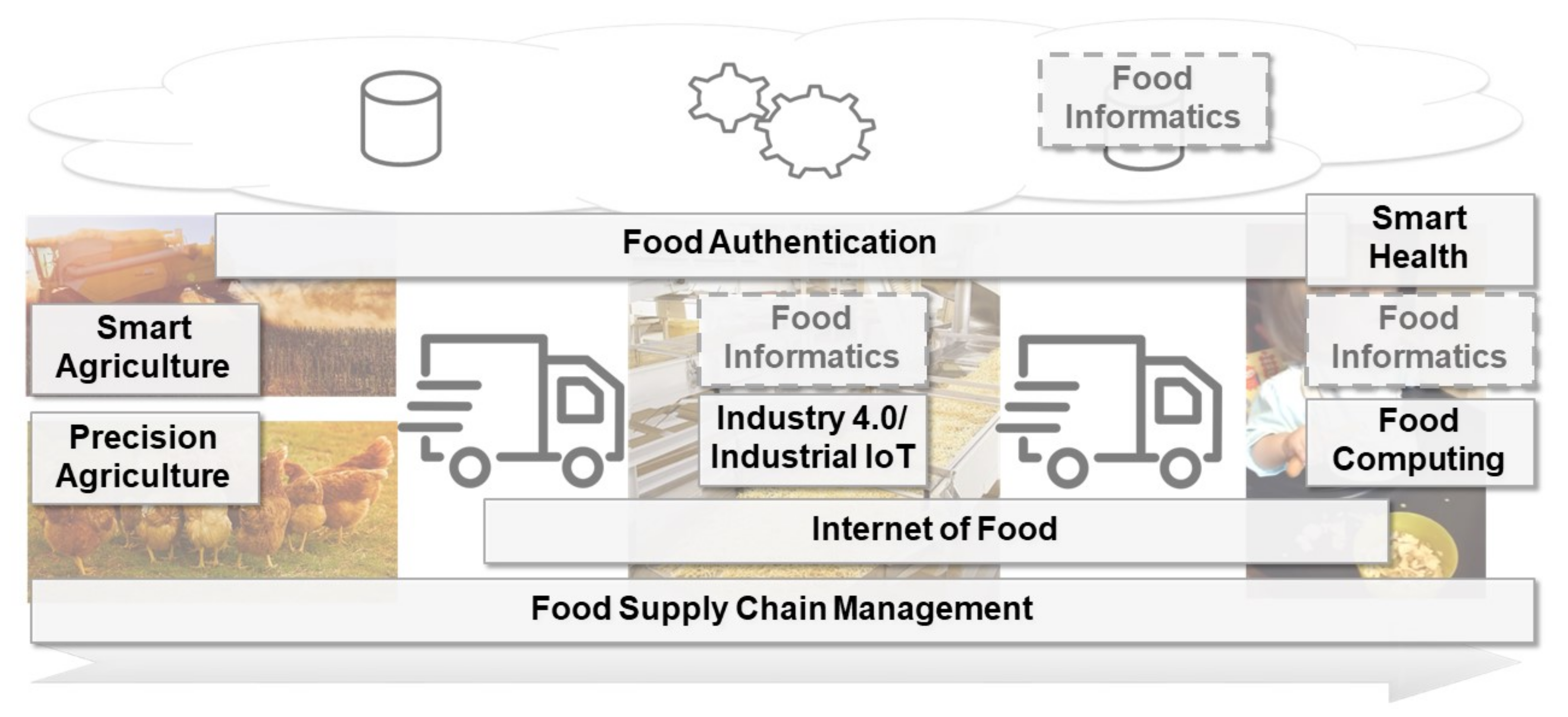
The increasing population of humans and their changing food consumption behavior, as well as the recent developments in the awareness for food sustainability, lead to new challenges for the production of food. Advances in the [[internet of things]] (IoT) and [[artificial intelligence]] (AI) technology, including [[machine learning]] and [[Data analysis|data analytics]], might help to account for these challenges. Several research perspectives—among them precision agriculture, industrial IoT, internet of food, and smart health—already provide new opportunities through digitalization. In this paper, we review the current state-of-the-art of the mentioned concepts. An additional concept to address is food informatics, which so far is mostly recognized as a mainly data-driven approach to support the production of food. In this review paper, we propose and discuss a new perspective for the concept of food informatics as a supportive discipline ... ('''[[Journal:Food informatics: Review of the current state-of-the-art, revised definition, and classification into the research landscape|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 17–23:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Kohn LearnHlthSys2022 6-1.jpg|240px]]</div> | |||
'''"[[Journal:Creating learning health systems and the emerging role of biomedical informatics|Creating learning health systems and the emerging role of biomedical informatics]]"''' | |||
The nature of [[information]] used in medicine has changed. In the past, we were limited to routine clinical data and published clinical trials. Today, we deal with massive, multiple data streams and easy access to new tests, ideas, and capabilities to process them. Whereas in the past getting information for decision-making was a challenge, today's clinicians have readily available access to information and data through the multitude of data-collecting devices, though it remains a challenge at times to analyze, evaluate, and prioritize it. As such, clinicians must become adept with the tools needed to deal with the era of big data, requiring a major change in how we learn to make decisions. Major change is often met with resistance and questions about value. A "learning health system" (LHS) is an enabler to encourage the development of such tools and demonstrate value in improved decision-making ... ('''[[Journal:Creating learning health systems and the emerging role of biomedical informatics|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 10–16:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Liscouski PlanDisruptLabOper2022.png|240px]]</div> | |||
'''"[[LII:Planning for Disruptions in Laboratory Operations|Planning for Disruptions in Laboratory Operations]]"''' | |||
A high-level of productivity is something laboratory management wants and those working for them strive to achieve. However, what happens when reality trips us up? We found out when [[COVID-19]] appeared. This work from laboratory informatics veteran Joe Liscouski examines how [[laboratory]] operations can be organized to meet that disruption, as well as other disruptions we may have to face. Many of these changes, including the introduction of new technologies and changing attitudes about work, were in the making already but at a much slower pace. Over the years, productivity has had many measures, from 40 to 60 hour work weeks and piece-work to pounds of material processed to samples run, all of which comes from a manufacturing mind set. People went to work in an office, lab, or production site, did their work, put in their time, and went home. That was in the timeframe leading up to the 1950s and '60s. Today, in 2022, things have changed ... ('''[[LII:Planning for Disruptions in Laboratory Operations|Full article...]]''')<br /> | |||
|- | |||
|<br /><<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 3–9:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Eiroa InsightsIntoImaging22 13.png|240px]]</div> | |||
'''"[[Journal:The current state of knowledge on imaging informatics: A survey among Spanish radiologists|The current state of knowledge on imaging informatics: A survey among Spanish radiologists]]"''' | |||
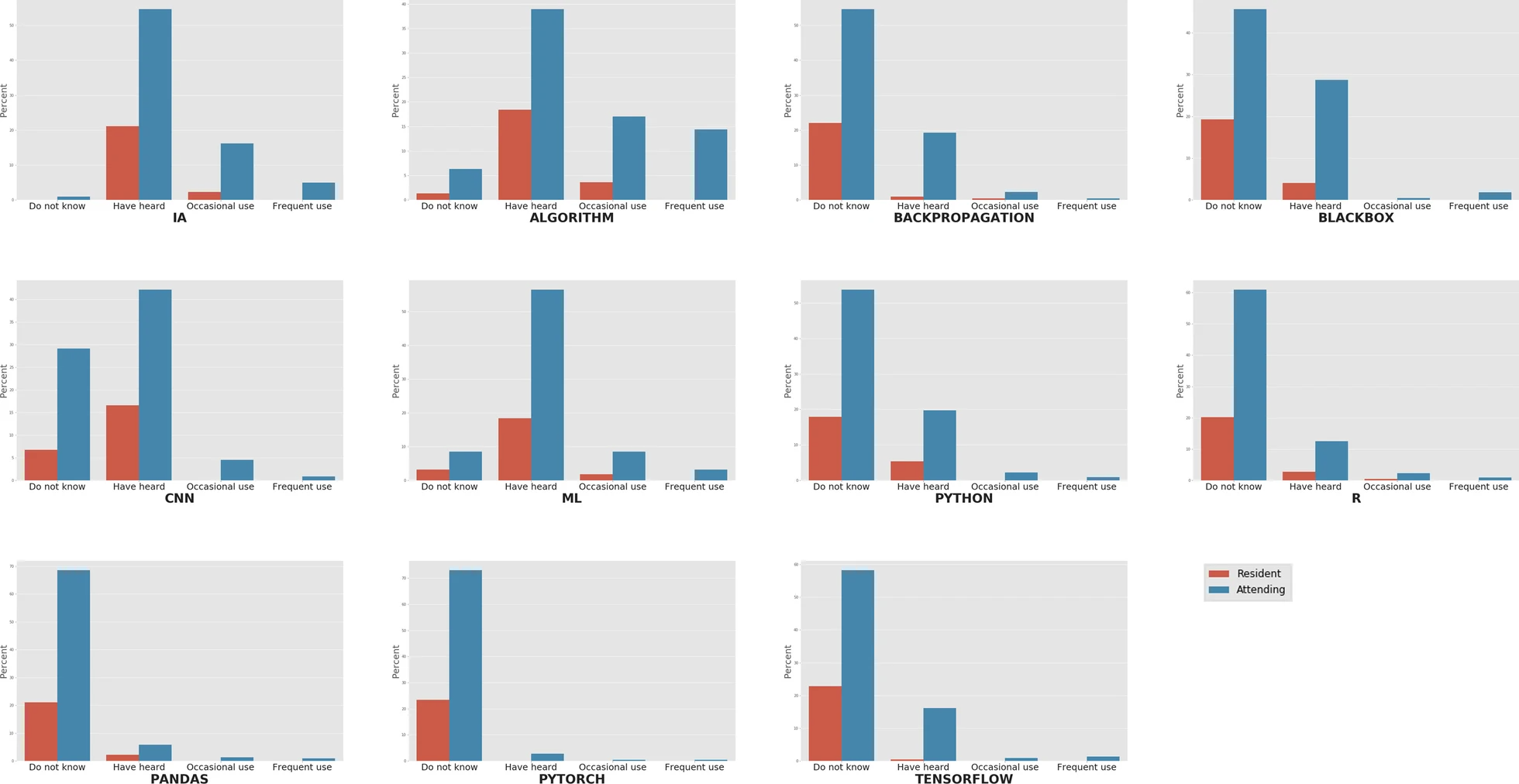
There is growing concern about the impact of [[artificial intelligence]] (AI) on radiology and the future of the profession. The aim of this study is to evaluate general knowledge and concerns about trends on [[imaging informatics]] among radiologists working in Spain (residents and attending physicians). For this purpose, an online survey among radiologists working in Spain was conducted with questions related to knowledge about terminology and technologies, need for a regulated academic training on AI, and concerns about the implications of the use of these technologies. A total of 223 radiologists answered the survey, of whom 23.3% were residents and 76.7% were attending physicians. General terms such as "AI" and "algorithm" had been heard of or read in at least 75.8% and 57.4% of the cases, respectively, while more specific terms were scarcely known. All the respondents considered that they should pursue academic training in [[medical informatics]] and new technologies, and 92.9% of them reckoned this preparation should be incorporated in the training program of the specialty ... ('''[[Journal:The current state of knowledge on imaging informatics: A survey among Spanish radiologists|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 26–October 02:</h2> | |||
'''"[[Journal:Emerging cybersecurity threats in radiation oncology|Emerging cybersecurity threats in radiation oncology]]"''' | |||
Modern image-guided radiation therapy is dependent on information technology and data storage applications that, like any other digital technology, are at risk from cyberattacks. Owing to a recent escalation in cyberattacks affecting radiation therapy treatments, the American Society for Radiation Oncology's ''Advances in Radiation Oncology'' is inaugurating a new special manuscript category devoted to [[cybersecurity]] issues. We conducted a review of emerging cybersecurity threats and a literature review of cyberattacks that affected [[Radiation oncologist|radiation oncology]] practices. In the last 10 years, numerous attacks have led to an interruption of radiation therapy for thousands of patients, and some of these catastrophic incidents have been described as being worse than [[coronavirus disease 2019]]'s impact on healthcare centers in New Zealand ... ('''[[Journal:Emerging cybersecurity threats in radiation oncology|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 19–25:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig7 Maury FrontDigHlth2021 3.jpg|240px]]</div> | |||
'''"[[Journal:An automated dashboard to improve laboratory COVID-19 diagnostics management|An automated dashboard to improve laboratory COVID-19 diagnostics management]]"''' | |||
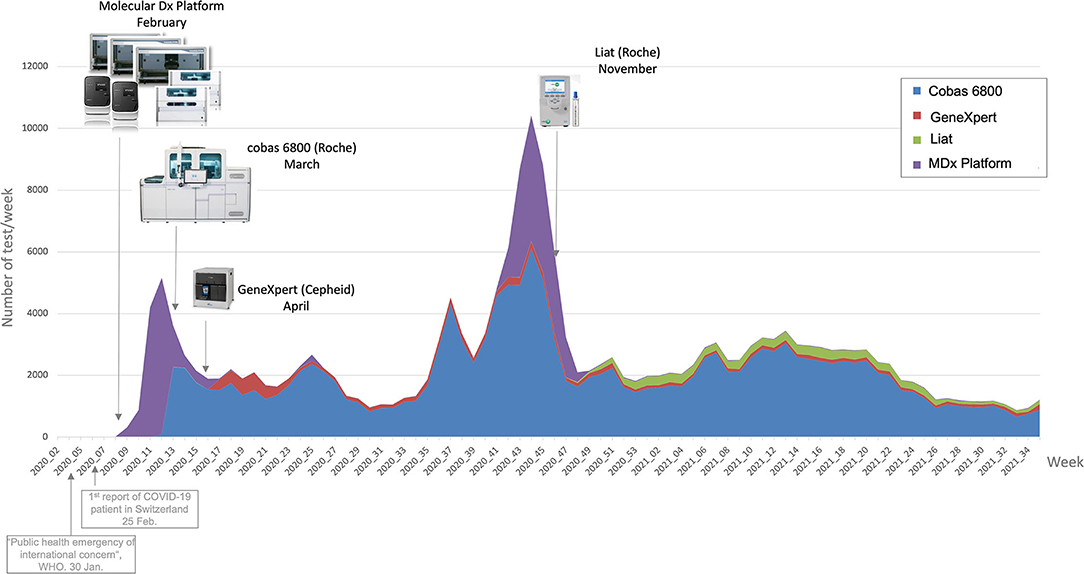
In response to the [[COVID-19]] [[pandemic]], our microbial diagnostic [[laboratory]] located in a university [[hospital]] has implemented several distinct [[SARS-CoV-2]] [[reverse transcription polymerase chain reaction]] (RT-PCR) systems in a very short time. More than 148,000 tests have been performed over 12 months, which represents about 405 tests per day, with peaks to more than 1,500 tests per days during the second wave. This was only possible thanks to [[Laboratory automation|automation]] and digitalization, to allow high-throughput, acceptable time to results and to maintain test reliability. An automated dashboard was developed to give access to key performance indicators (KPIs) to improve laboratory operational management. RT-PCR data extraction of four respiratory viruses—SARS-CoV-2, influenza A and B, and RSV—from our [[laboratory information system]] (LIS) was automated. This included ... ('''[[Journal:An automated dashboard to improve laboratory COVID-19 diagnostics management|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 12–18:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Yeste AdvLabMed2021 2-3.jpg|240px]]</div> | |||
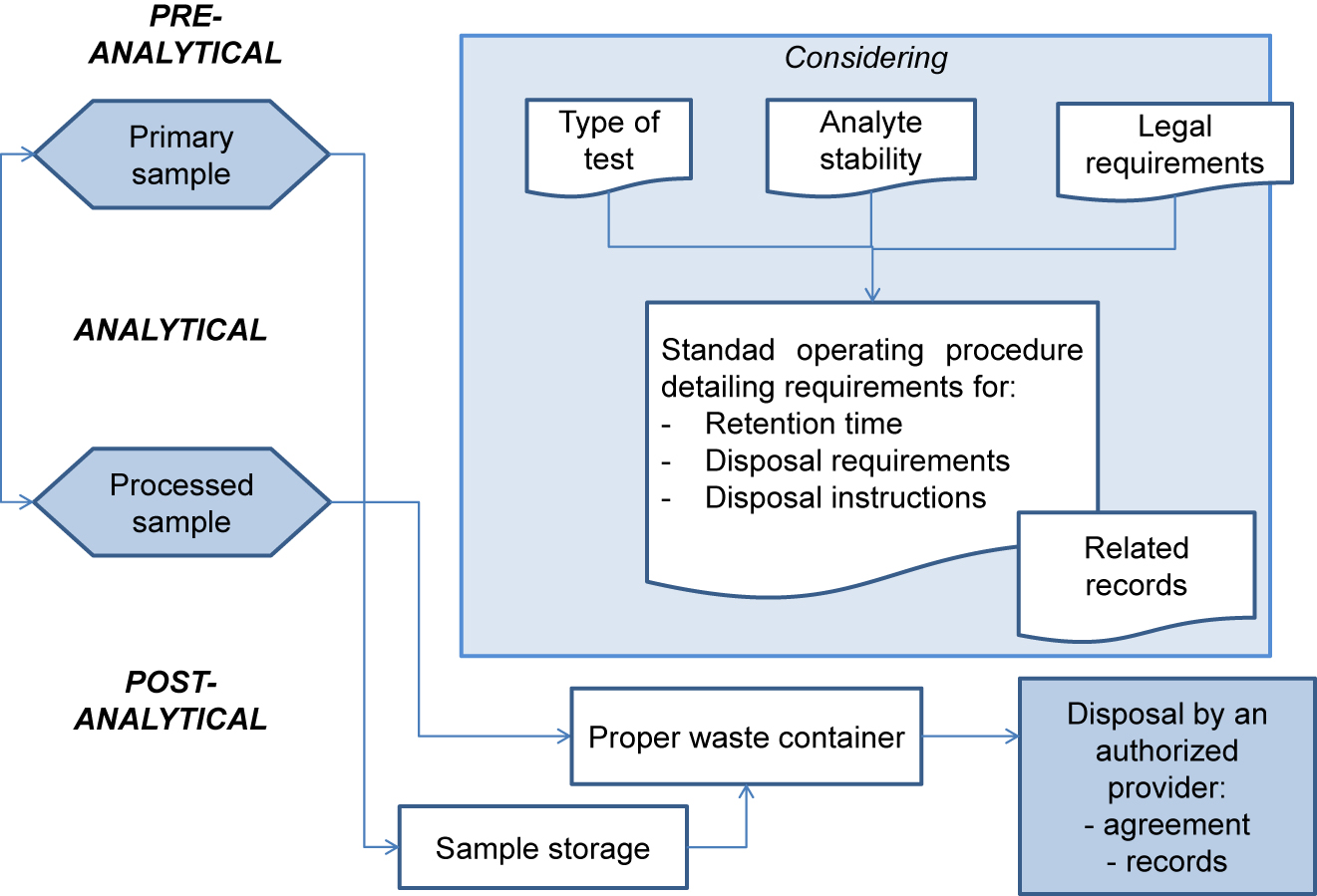
'''"[[Journal:Management of post-analytical processes in the clinical laboratory according to ISO 15189:2012: Considerations about the management of clinical samples, ensuring quality of post-analytical processes and laboratory information management|Management of post-analytical processes in the clinical laboratory according to ISO 15189:2012: Considerations about the management of clinical samples, ensuring quality of post-analytical processes and laboratory information management]]"''' | |||
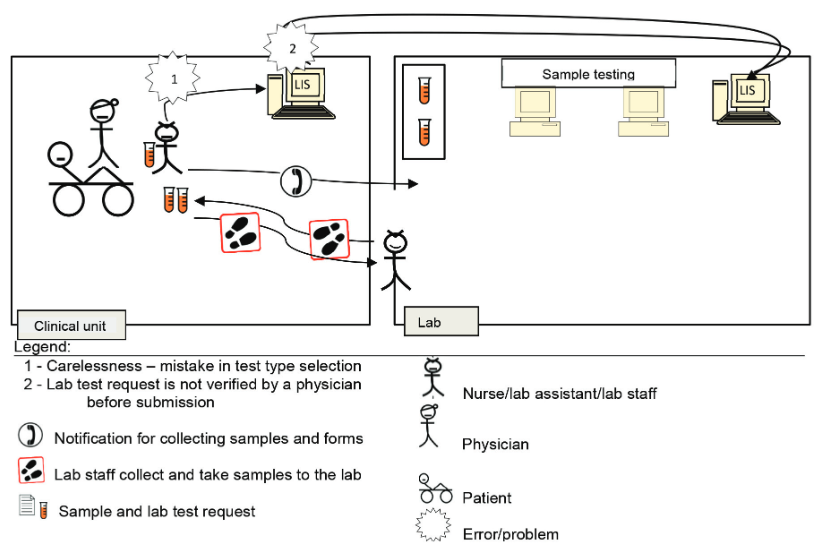
[[ISO 15189|ISO 15189:2012 ''Medical laboratories — Requirements for quality and competence'']] establishes the requirements for clinical specimen management, ensuring the [[Quality (business)|quality]] of processes and [[laboratory]] [[information management]]. ENAC (Entidad Nacional de Acreditación), the sole accreditation authority in Spain, established the requirements for the authorized use of the ISO 15189 accreditation label in reports issued by accredited laboratories. These recommendations are applicable to the lab's post-analytical processes and the professionals involved. The standard requires laboratories to define and document the duration and conditions of specimen [[Retention period|retention]]. Laboratories are also required to design an internal [[quality control]] scheme to verify whether post-analytical activities attain the expected standards. Information management requirements are also established, and laboratories are required to design a contingency plan to ensure the communication of laboratory results ... ('''[[Journal:Management of post-analytical processes in the clinical laboratory according to ISO 15189:2012: Considerations about the management of clinical samples, ensuring quality of post-analytical processes and laboratory information management|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 05–11:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig7 Fraggetta Diagnostics21 11-10.png|240px]]</div> | |||
'''"[[Journal:A survival guide for the rapid transition to a fully digital workflow: The Caltagirone example|A survival guide for the rapid transition to a fully digital workflow: The Caltagirone example]]"''' | |||
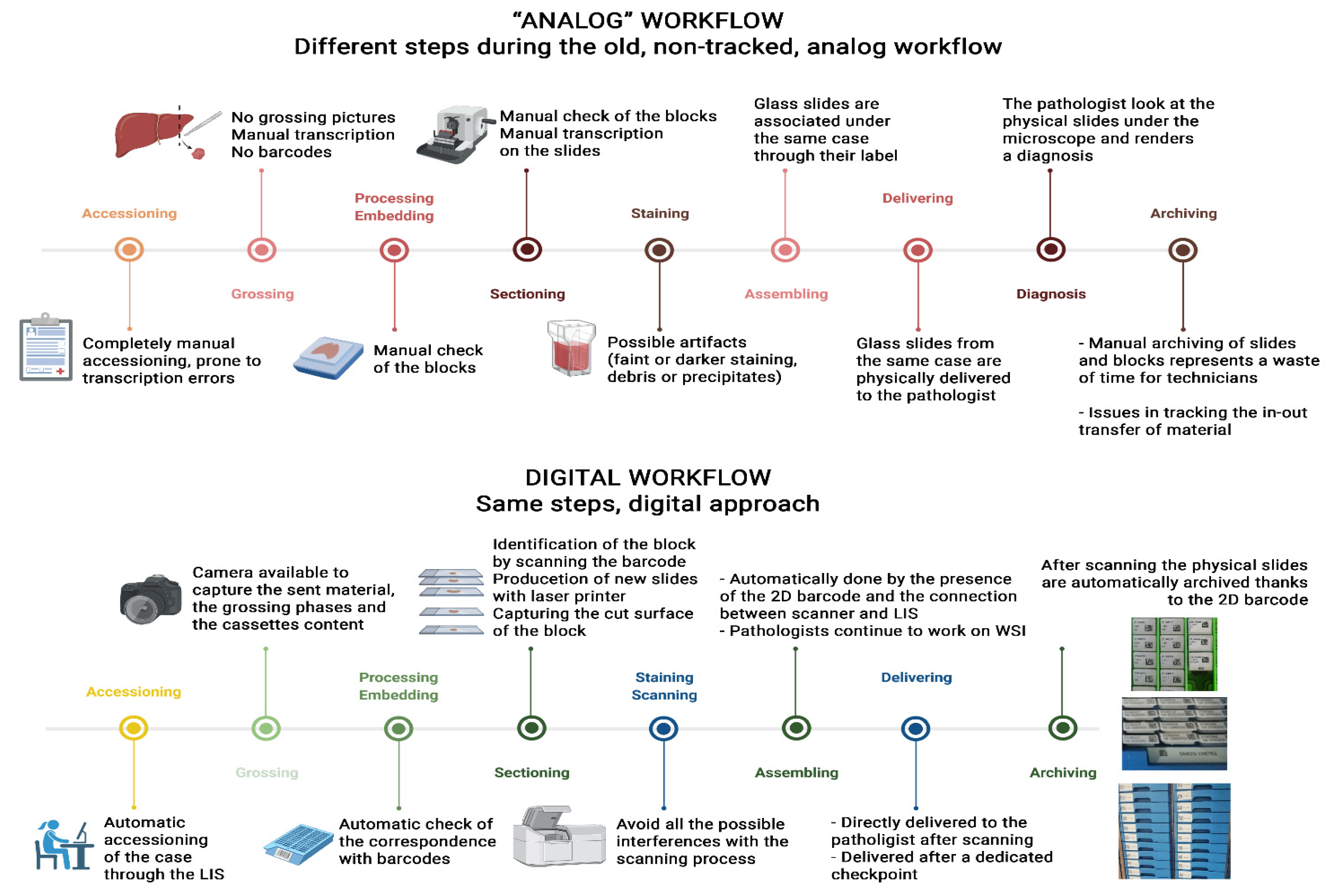
[[Digital pathology]] for the routine assessment of cases for primary diagnosis has been implemented by few [[Laboratory|laboratories]] worldwide. The Gravina Hospital in Caltagirone (Sicily, Italy), which collects cases from seven different [[hospital]]s distributed in the Catania area, converted its entire [[workflow]] to digital starting from 2019. Before the transition, the Caltagirone [[pathology]] laboratory was characterized by a non-tracked workflow, based on paper requests, and hand-written blocks and slides, as well as manual assembling and delivering of the cases and glass slides to the pathologists. Moreover, the arrangement of the spaces and offices in the department was illogical and under-productive for the linearity of the workflow. For these reasons, an adequate 2D [[barcode]] system for tracking purposes, the redistribution of laboratory spaces, and the implementation of whole-slide imaging (WSI) technology based on a [[laboratory information system]] (LIS)-centric approach ... ('''[[Journal:A survival guide for the rapid transition to a fully digital workflow: The Caltagirone example|Full article...]]''')<br /> | |||
|- | |||
|<br /><<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 29–September 04:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Asiimwe JofTransMed21 19.png|240px]]</div> | |||
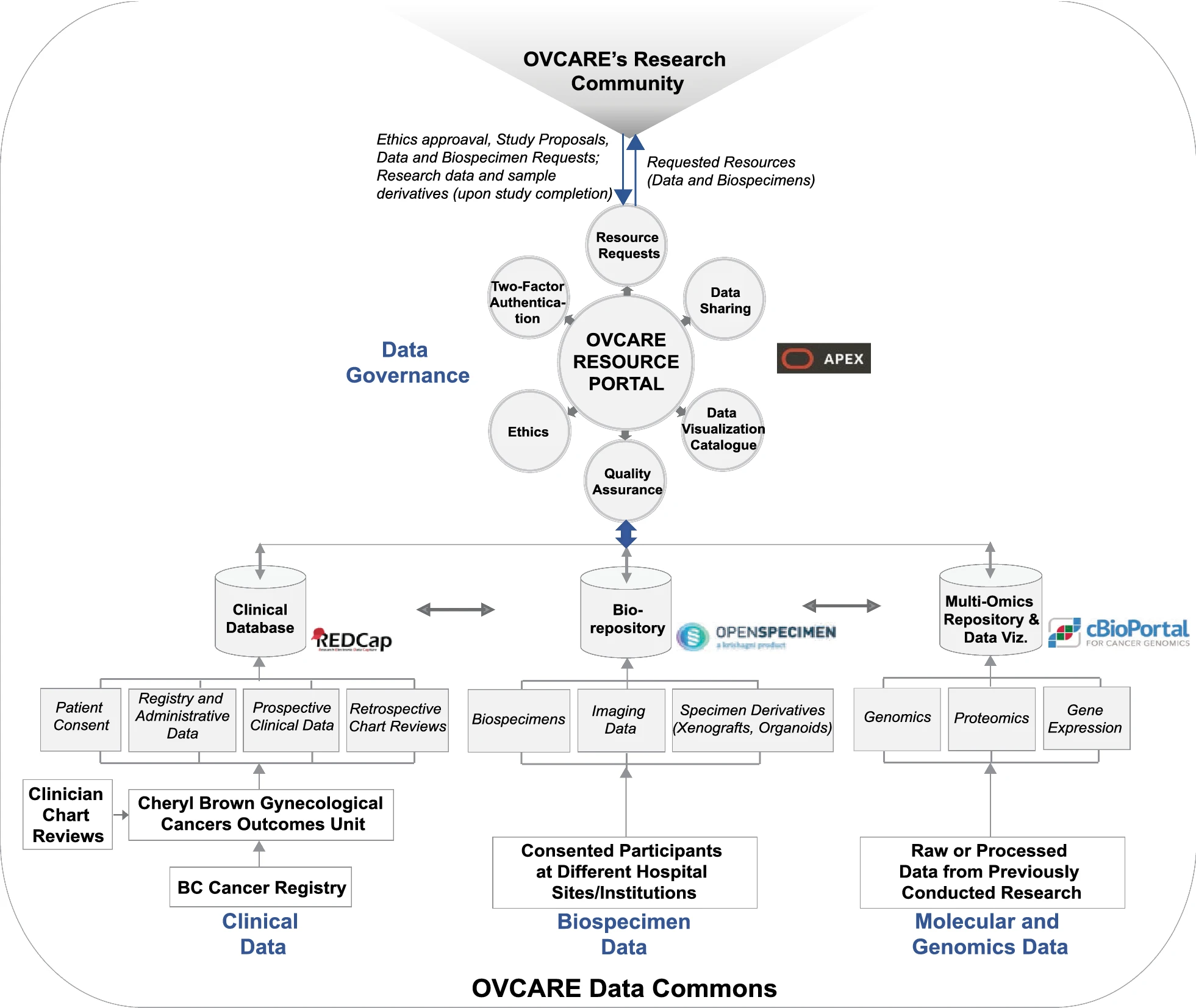
'''"[[Journal:From biobank and data silos into a data commons: Convergence to support translational medicine|From biobank and data silos into a data commons: Convergence to support translational medicine]]"''' | |||
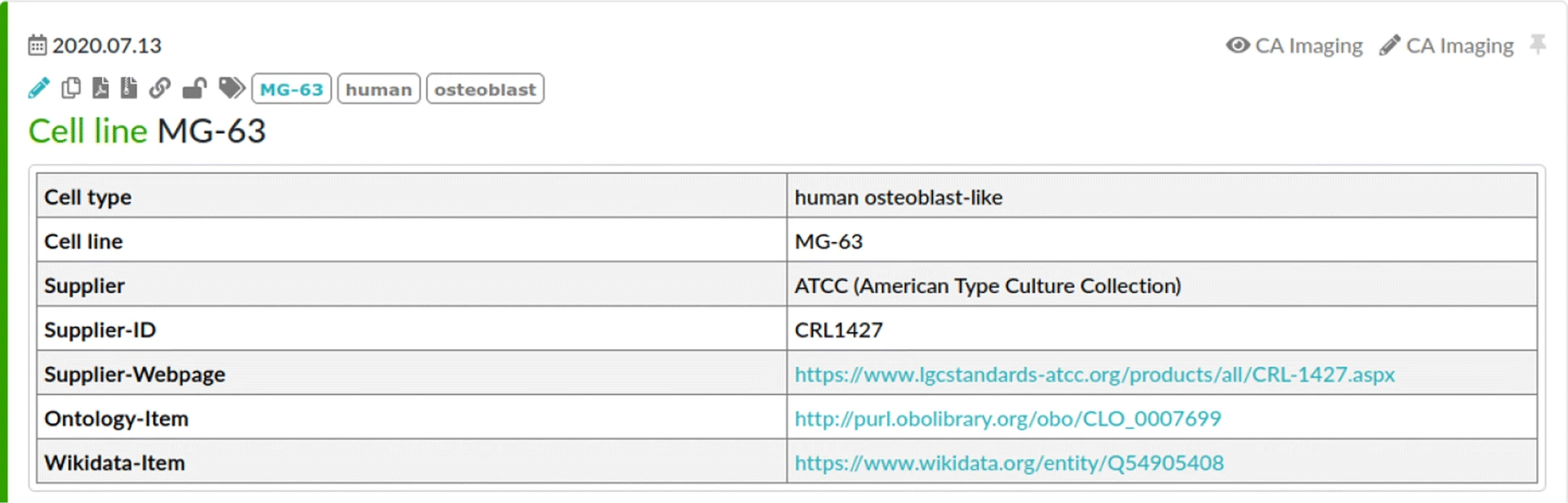
To drive [[Translational research|translational medicine]], modern day [[biobank]]s need to integrate with other sources of data (e.g., [[Health informatics|clinical]], [[genomics]]) to support novel data-intensive research. Currently, vast amounts of research and clinical data remain in silos, held and managed by individual researchers, operating under different standards and governance structures; such a framework impedes sharing and effective use of data. In this article, we describe the journey of British Columbia’s Gynecological Cancer Research Program (OVCARE) in moving a traditional tumor biobank, outcomes unit, and a collection of data silos into an integrated [[Open data#Policies and strategies|data commons]] to support data standardization and [[Data sharing|resource sharing]] under collaborative governance, as a means of providing the gynecologic cancer research community in British Columbia access to tissue samples and associated clinical and [[Molecular diagnostics|molecular]] data from thousands of patients ... ('''[[Journal:From biobank and data silos into a data commons: Convergence to support translational medicine|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 15–21:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Kachuwaire OneHealth2021 13.jpg|240px]]</div> | |||
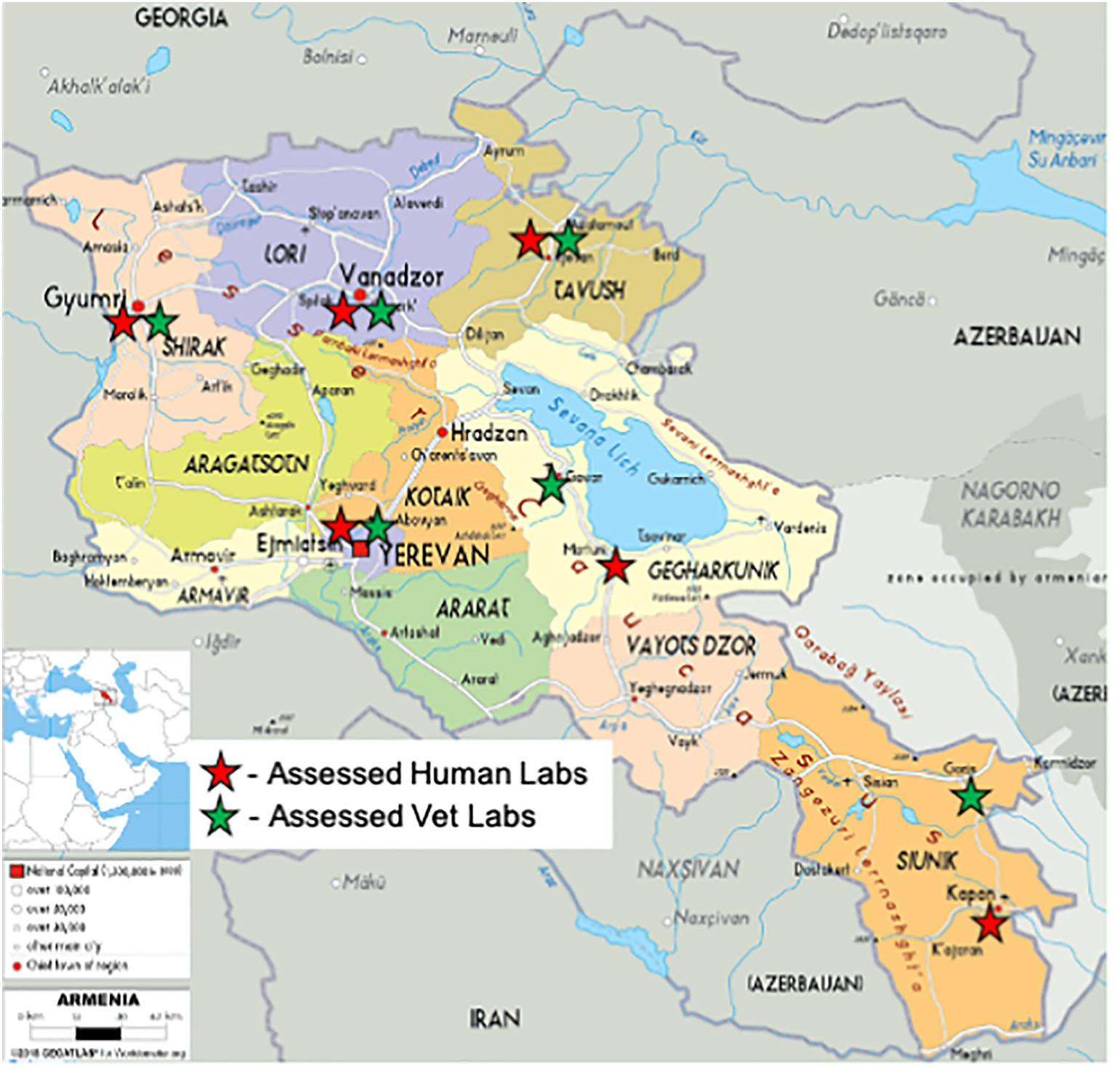
'''"[[Journal:Quality management system implementation in human and animal laboratories|Quality management system implementation in human and animal laboratories]]"''' | |||
The ability to rapidly detect emerging and re-emerging threats relies on a strong network of [[Laboratory|laboratories]] providing high-quality testing services. Improving laboratory [[Quality management system|quality management systems]] (QMS) to ensure that these laboratories effectively play their critical role using a tailored stepwise approach can assist them to comply with World Health Organization's (WHO) International Health Regulations (IHRs), as well as the World Organization for Animal Health's (OIE) guidelines. Fifteen (15) laboratories in Armenia's human and veterinary laboratory networks were enrolled into a QMS strengthening program from 2017 to 2020. Training was provided for key staff, resulting in an implementation plan developed to address gaps. Routine mentorship visits were conducted. Audits were undertaken at baseline and post-implementation using standardized checklists to assess laboratory improvements ... ('''[[Journal:Quality management system implementation in human and animal laboratories|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 08–14:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Krantz FrontPlantSci2021 12.jpg|240px]]</div> | |||
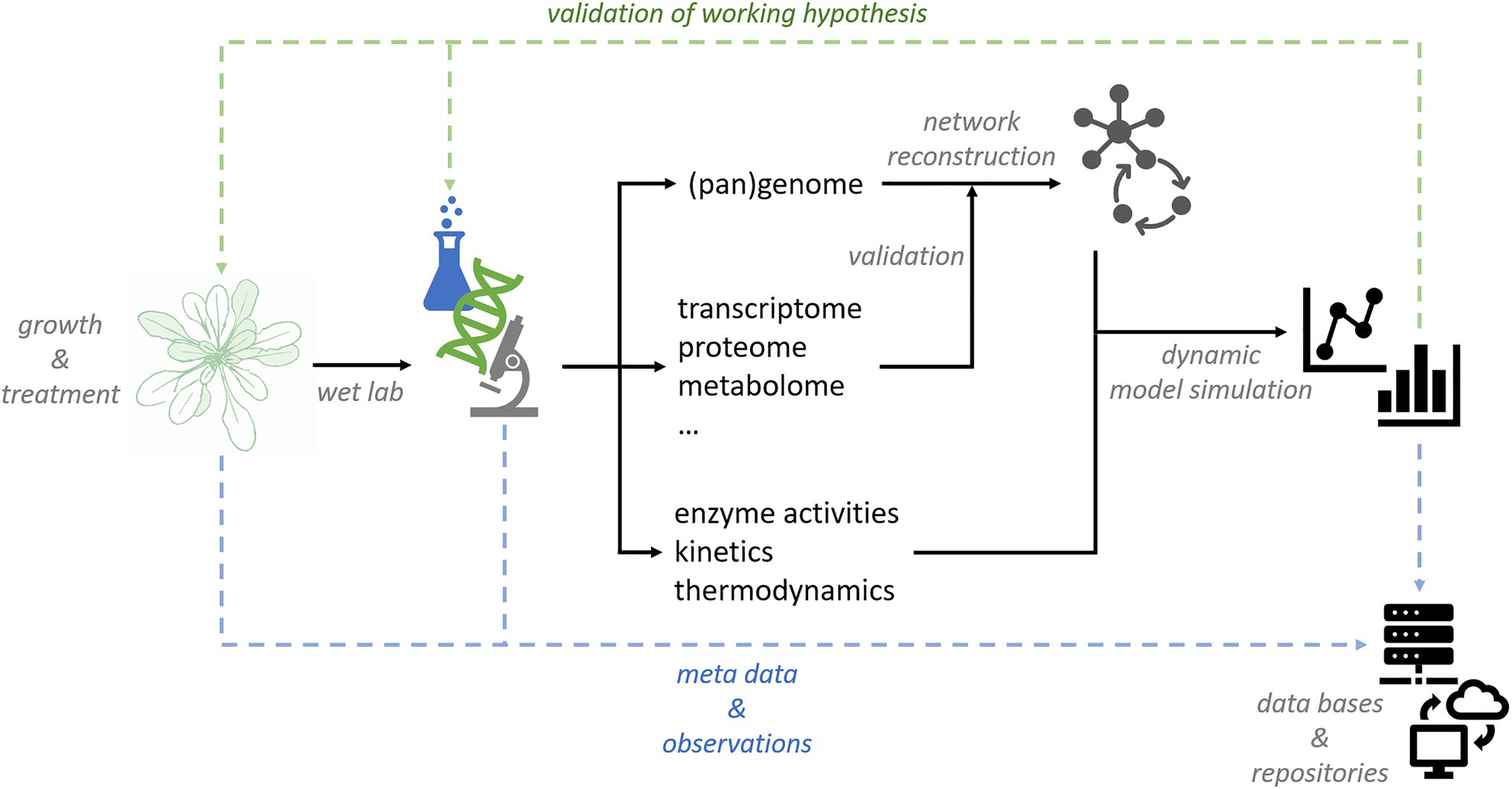
'''"[[Journal:Data management and modeling in plant biology|Data management and modeling in plant biology]]"''' | |||
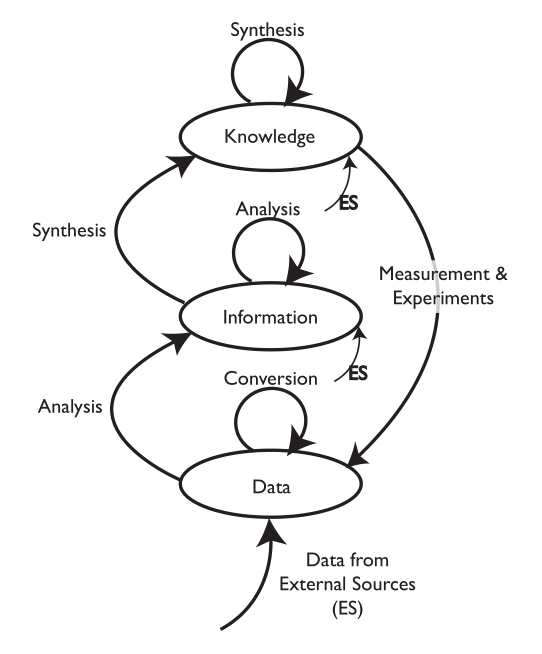
The study of plant-environment interactions is a multidisciplinary research field. With the emergence of quantitative large-scale and high-throughput techniques, the amount and dimensionality of experimental data have strongly increased. Appropriate strategies for data storage, [[Information management|management]], and [[Data analysis|evaluation]] are needed to make efficient use of experimental findings. Computational approaches to [[data mining]] are essential for deriving statistical trends and signatures contained in data matrices. Although, current biology is challenged by high data dimensionality in general, this is particularly true for plant biology. As sessile organisms, plants have to cope with environmental fluctuations ... ('''[[Journal:Data management and modeling in plant biology|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 01-07:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Damerow DataSciJourn21 20-1.png|240px]]</div> | |||
'''"[[Journal:Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences|Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences]]"''' | |||
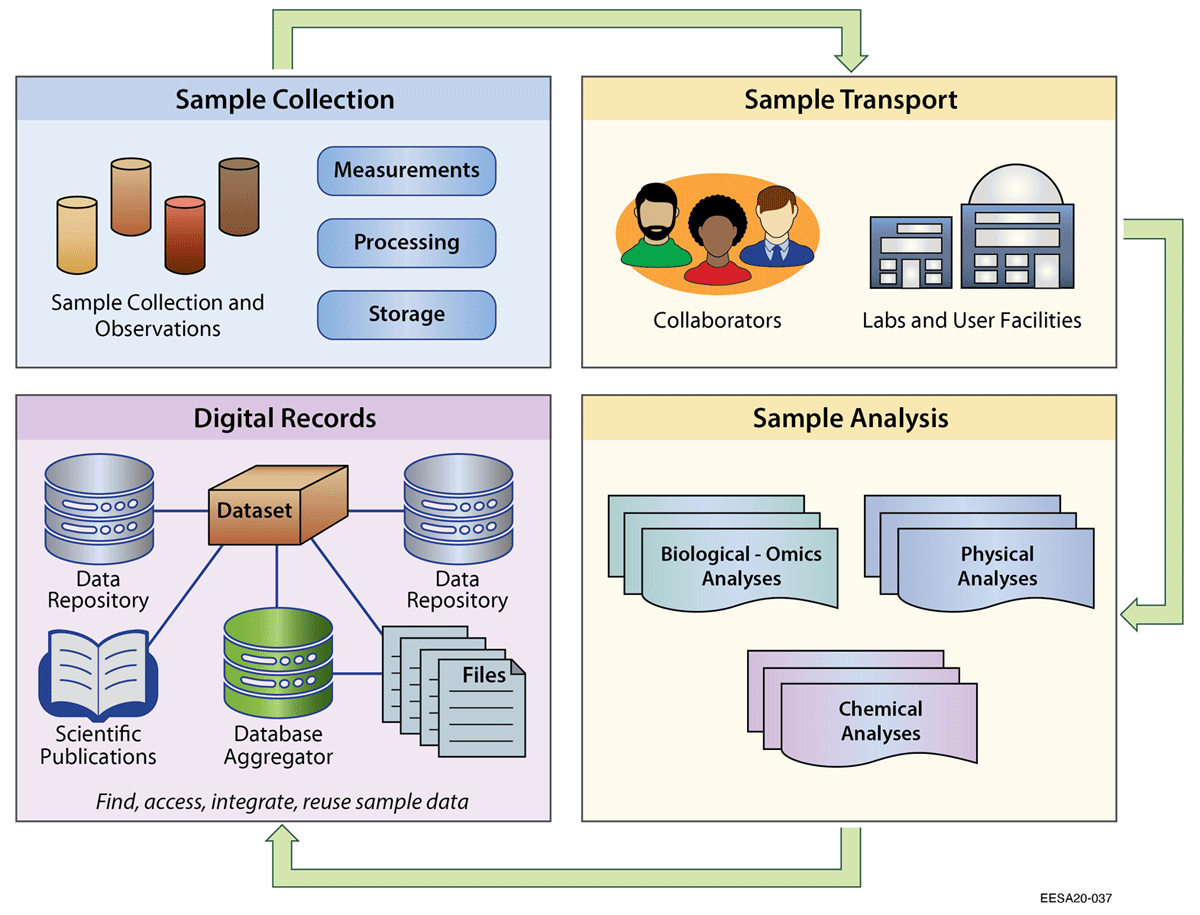
Physical [[Sample (material)|samples]] are foundational entities for research across the biological, Earth, and environmental sciences. Data generated from sample-based analyses are not only the basis of individual studies, but can also be integrated with other data to answer new and broader-scale questions. Ecosystem studies increasingly rely on multidisciplinary team-based science to study climate and environmental changes. While there are widely adopted conventions within certain domains to describe sample data, these have gaps when applied in a multidisciplinary context. In this study, we reviewed existing practices for identifying, characterizing, and linking related environmental samples. We then tested practicalities of assigning persistent identifiers to samples, with standardized [[metadata]], in a pilot field test involving eight United States Department of Energy projects. ... ('''[[Journal:Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 25–31:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Fraggetta Diagnostics21 11-11.png|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Fraggetta Diagnostics21 11-11.png|240px]]</div> | ||
'''"[[Journal:Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of Digital and Integrative Pathology (ESDIP)|Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of Digital and Integrative Pathology (ESDIP)]]"''' | '''"[[Journal:Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of Digital and Integrative Pathology (ESDIP)|Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of Digital and Integrative Pathology (ESDIP)]]"''' | ||
| Line 159: | Line 278: | ||
'''"[[Journal:Informatics-driven quality improvement in the modern histology lab|Informatics-driven quality improvement in the modern histology lab]]"''' | '''"[[Journal:Informatics-driven quality improvement in the modern histology lab|Informatics-driven quality improvement in the modern histology lab]]"''' | ||
[[Laboratory information system]]s (LISs) and [[data visualization]] techniques have untapped potential in [[Anatomical pathology|anatomic pathology]] [[Laboratory|laboratories]]. The pre-built functionalities of an LIS do not address all the needs of a modern [[histology]] laboratory. For instance, “go live” is not the end of LIS customization, only the beginning. After closely evaluating various histology lab [[workflows]], we implemented several custom [[Data analysis|data analytics]] dashboards and additional LIS functionalities to monitor and address weaknesses. Herein, we present our experience with LIS and data-tracking solutions that improved trainee education, slide logistics, staffing and instrumentation lobbying, and task tracking. The latter was addressed through the creation of a novel “status board” akin to those seen in inpatient wards. These use-cases can benefit other histology laboratories. ('''[[Journal:Informatics-driven quality improvement in the modern histology lab|Full article...]]''')<br /> | [[Laboratory information system]]s (LISs) and [[data visualization]] techniques have untapped potential in [[Anatomical pathology|anatomic pathology]] [[Laboratory|laboratories]]. The pre-built functionalities of an LIS do not address all the needs of a modern [[histology]] laboratory. For instance, “go live” is not the end of LIS customization, only the beginning. After closely evaluating various histology lab [[Workflow|workflows]], we implemented several custom [[Data analysis|data analytics]] dashboards and additional LIS functionalities to monitor and address weaknesses. Herein, we present our experience with LIS and data-tracking solutions that improved trainee education, slide logistics, staffing and instrumentation lobbying, and task tracking. The latter was addressed through the creation of a novel “status board” akin to those seen in inpatient wards. These use-cases can benefit other histology laboratories. ('''[[Journal:Informatics-driven quality improvement in the modern histology lab|Full article...]]''')<br /> | ||
|- | |- | ||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 07–13:</h2> | |<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 07–13:</h2> | ||
Latest revision as of 23:51, 11 April 2024
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 - 2023 - 2024 |
Featured article of the week archive - 2022
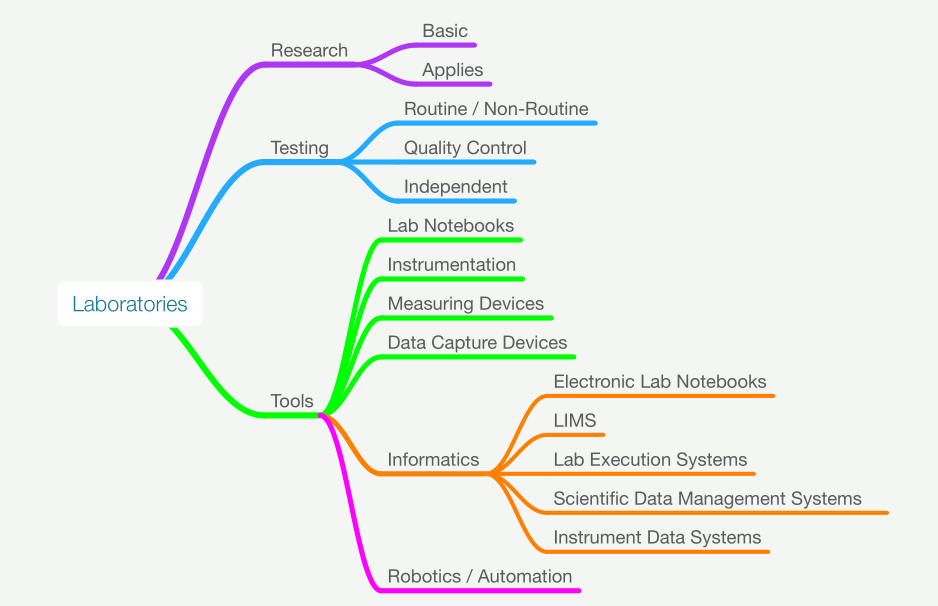
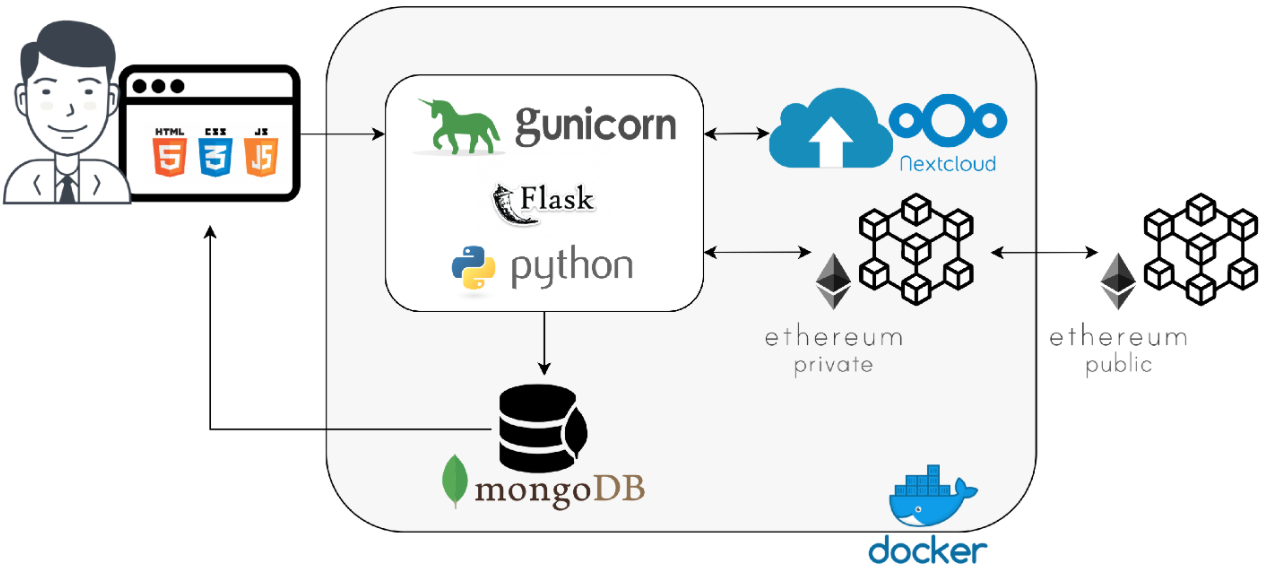
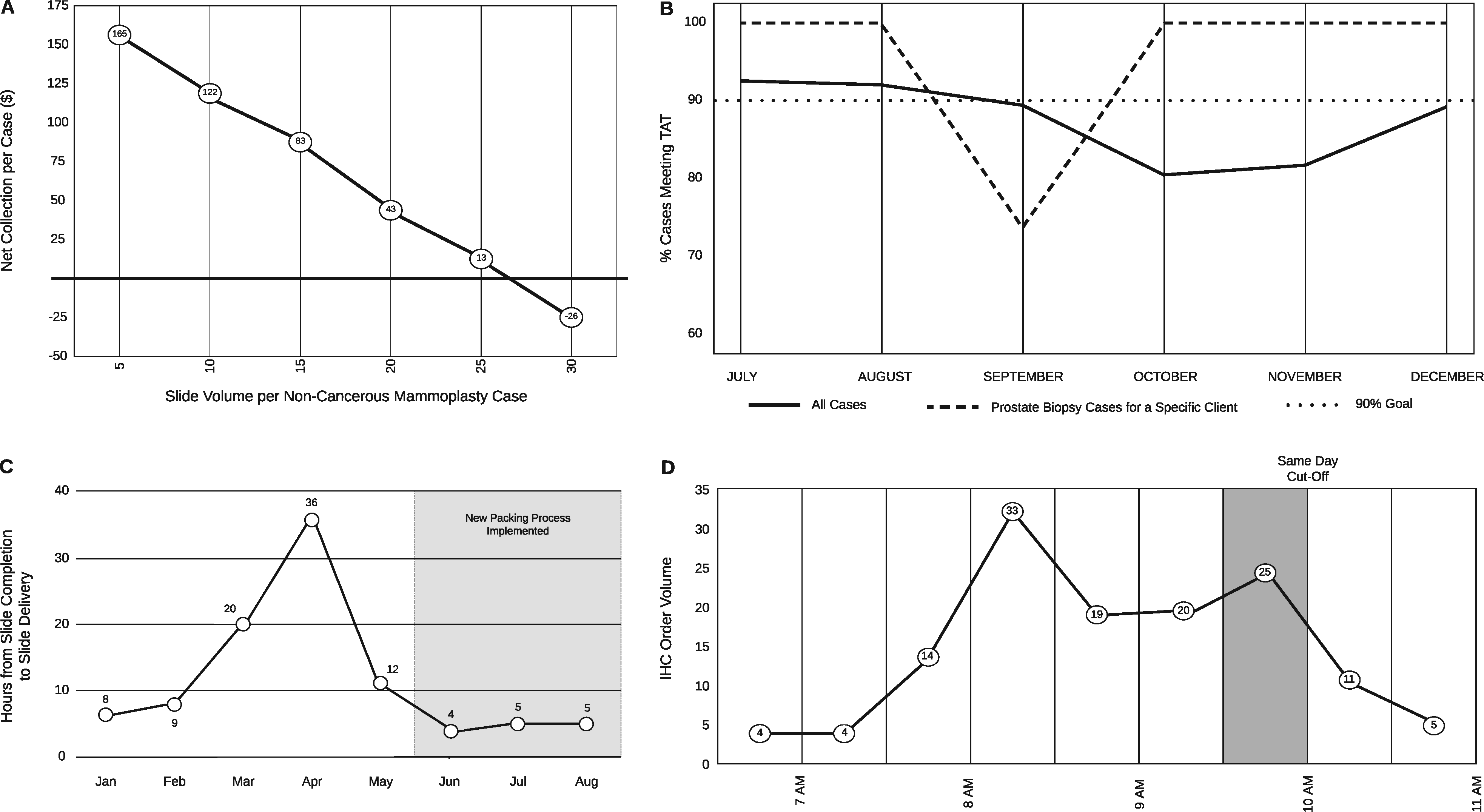
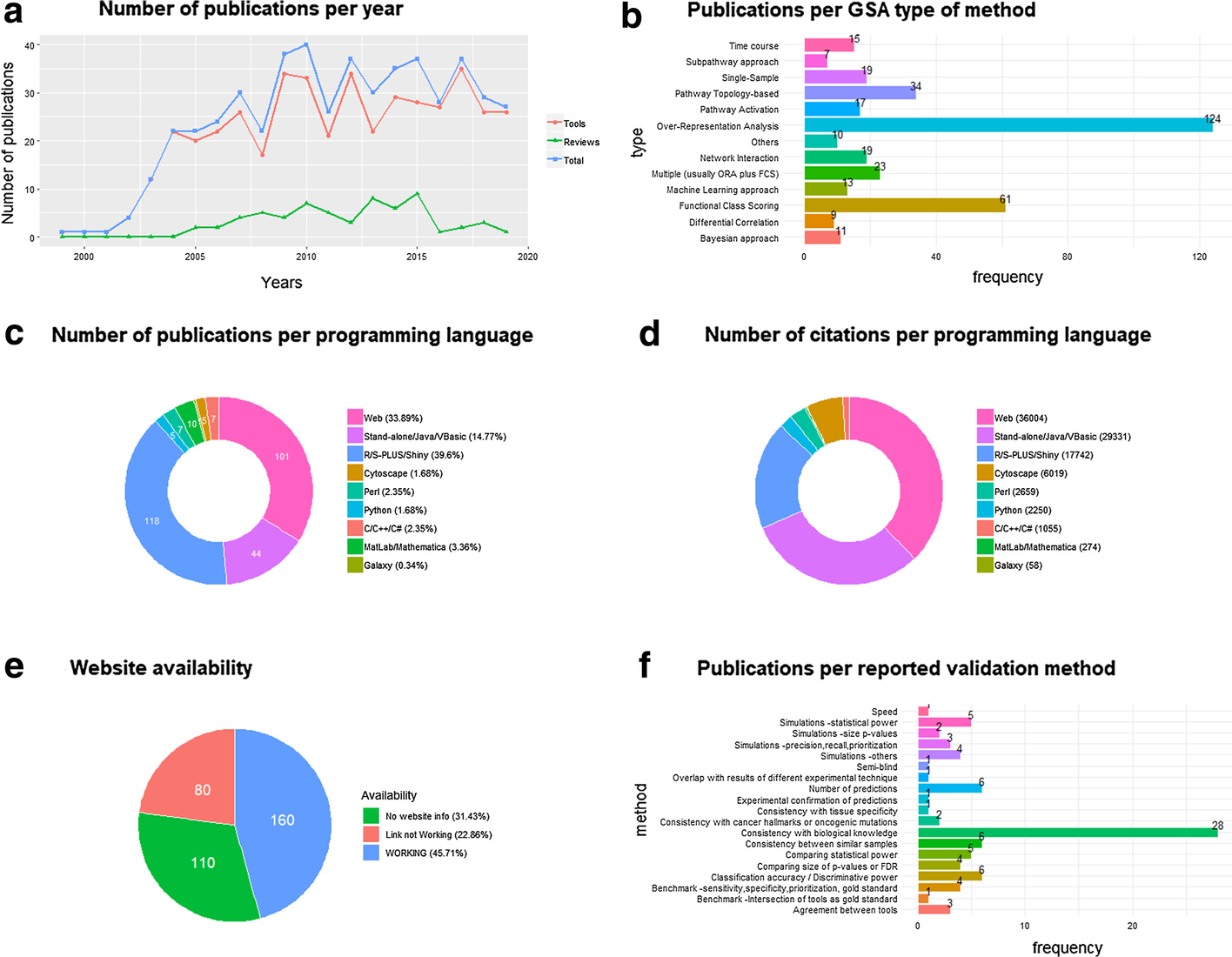
Welcome to the LIMSwiki 2022 archive for the Featured Article of the Week.
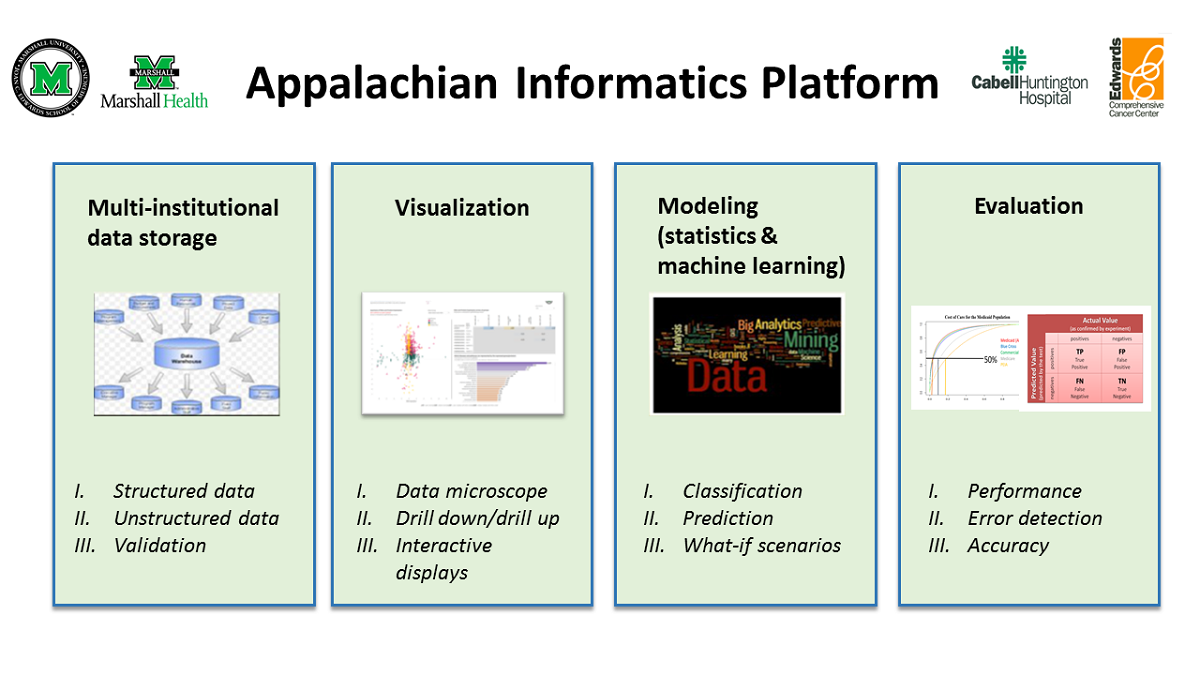
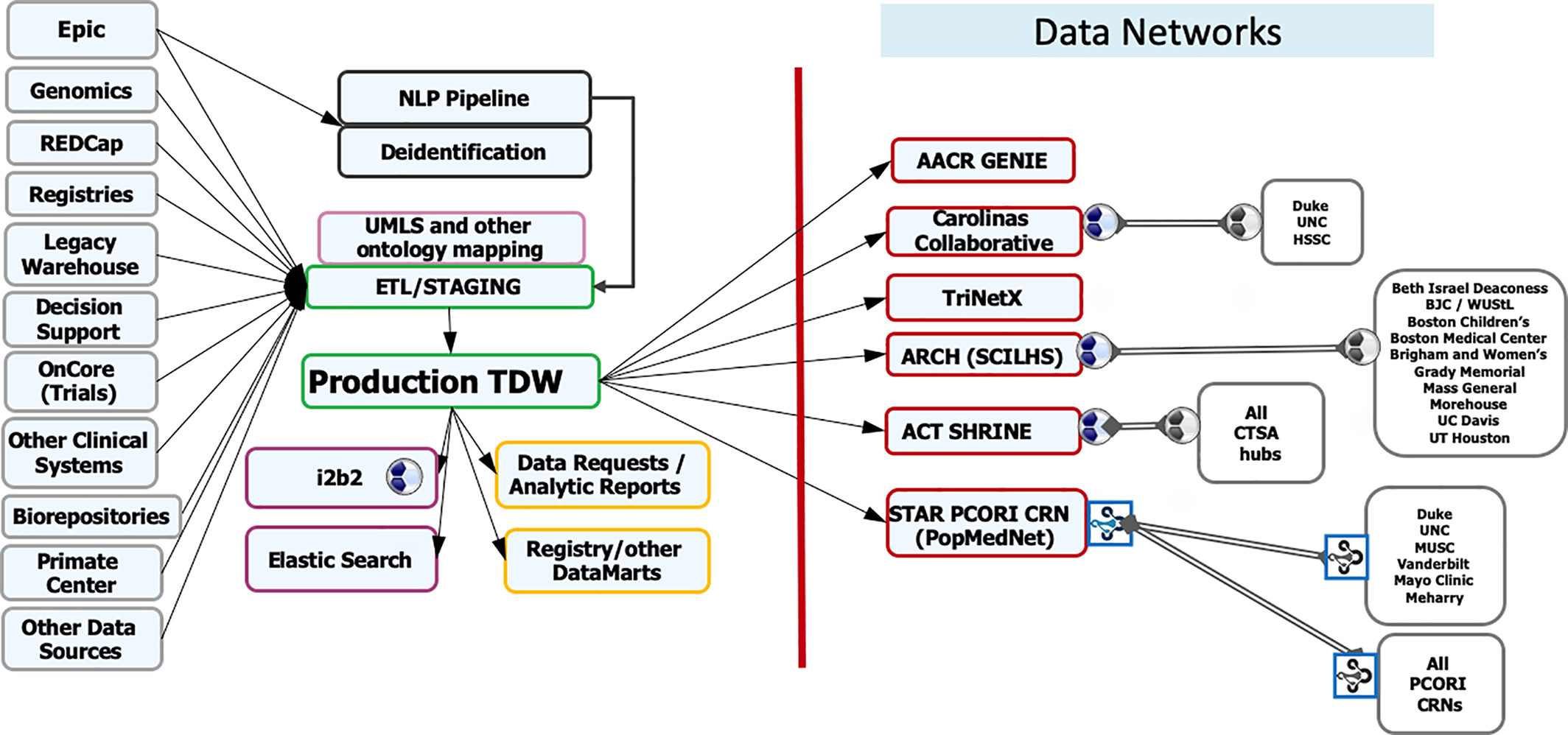
Featured article of the week: December 19–25:"Fueling clinical and translational research in Appalachia: Informatics platform approach" The Appalachian population is distinct, not just culturally and geographically but also in its healthcare needs, facing the most health care disparities in the United States. To meet these unique demands, Appalachian medical centers need an arsenal of analytics and data science tools with the foundation of a centralized data warehouse to transform healthcare data into actionable clinical interventions. However, this is an especially challenging task given the fragmented state of medical data within Appalachia and the need for integration of other types of data such as environmental, social, and economic with medical data. This paper aims to present the structure and process of the development of an integrated platform at a midlevel Appalachian academic medical center, along with its initial uses ... (Full article...)
|