Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text) |
Shawndouglas (talk | contribs) (Updated article of the week text) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:GA Karim JofKSUScience2022 34-2.jpg|240px]]</div> | ||
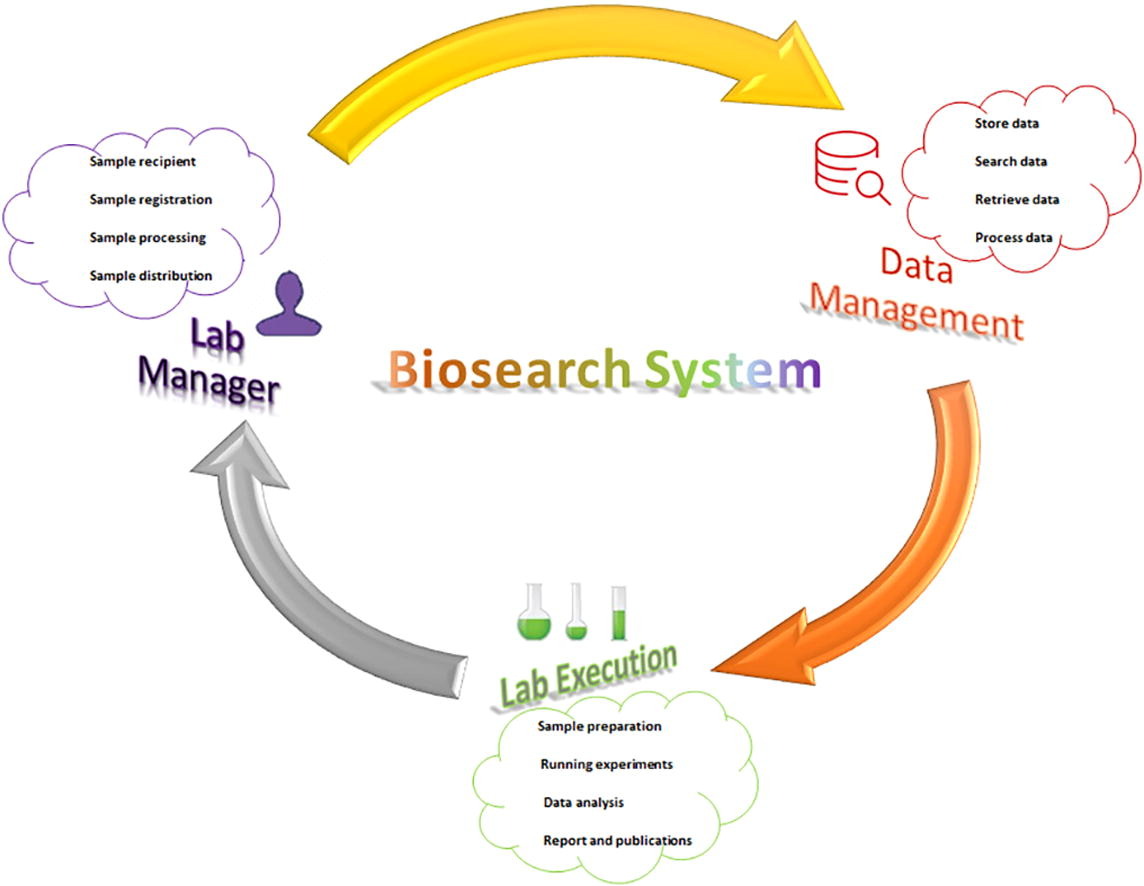
'''"[[Journal: | '''"[[Journal:Development of Biosearch System for biobank management and storage of disease-associated genetic information|Development of Biosearch System for biobank management and storage of disease-associated genetic information]]"''' | ||
Databases and software are important to manage modern high-throughput [[Laboratory|laboratories]] and store clinical and [[Genome informatics|genomic information]] for [[quality assurance]]. Commercial software is expensive, with proprietary code issues, while academic versions have adaptation issues. Our aim was to develop an adaptable in-house software system that can store specimen- and disease-associated genetic information in [[biobank]]s to facilitate [[translational research]]. A prototype was designed per the research requirements, and computational tools were used to develop the software under three tiers, using Visual Basic and ASP.net for the presentation tier, SQL Server for the data tier, and Ajax and JavaScript for the business tier. We retrieved specimens from the biobank using this software and performed microarray-based transcriptomic analysis to detect differentially expressed genes (DEGs) ... ('''[[Journal:Development of Biosearch System for biobank management and storage of disease-associated genetic information|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
{{flowlist | | {{flowlist | | ||
* [[Journal:Establishing a common nutritional vocabulary: From food production to diet|Establishing a common nutritional vocabulary: From food production to diet]] | |||
* [[Journal:Designing a knowledge management system for naval materials failures|Designing a knowledge management system for naval materials failures]] | * [[Journal:Designing a knowledge management system for naval materials failures|Designing a knowledge management system for naval materials failures]] | ||
* [[Journal:Laboratory information management system for COVID-19 non-clinical efficacy trial data|Laboratory information management system for COVID-19 non-clinical efficacy trial data]] | * [[Journal:Laboratory information management system for COVID-19 non-clinical efficacy trial data|Laboratory information management system for COVID-19 non-clinical efficacy trial data]] | ||
}} | }} | ||
Revision as of 14:15, 13 March 2023
Databases and software are important to manage modern high-throughput laboratories and store clinical and genomic information for quality assurance. Commercial software is expensive, with proprietary code issues, while academic versions have adaptation issues. Our aim was to develop an adaptable in-house software system that can store specimen- and disease-associated genetic information in biobanks to facilitate translational research. A prototype was designed per the research requirements, and computational tools were used to develop the software under three tiers, using Visual Basic and ASP.net for the presentation tier, SQL Server for the data tier, and Ajax and JavaScript for the business tier. We retrieved specimens from the biobank using this software and performed microarray-based transcriptomic analysis to detect differentially expressed genes (DEGs) ... (Full article...)
Recently featured: