Journal:Establishing a common nutritional vocabulary: From food production to diet
| Full article title | Establishing a common nutritional vocabulary: From food production to diet |
|---|---|
| Journal | Frontiers in Nutrition |
| Author(s) | Andrés-Hernández, Liliana; Blumberg, Kai; Walls, Ramona L.; Dooley, Damion; Mauleon, Ramil; Lange, Matthew; Weber, Magalie; Chan, Lauren; Malik, Adnan; Møller, Anders; Ireland, Jayne; Segovia, Lucia; Zhang, Xuhuiqun; Burton-Freeman, Britt; Magelli, Paul; Schriever, Andrew; Forester, Shavawn M.; Liu, Lei; King, Graham J. |
| Author affiliation(s) | Southern Cross University, University of Arizona, Critical Path Institute, Simon Fraser University, IC-FOODS, INRAE BIA, Oregon State University, EMBL-EBI, Danish Food Informatics, University of London, Illinois Institute of Technology, WISEcode LLC, Nutrient Institute LLC, University of Nottingham |
| Primary contact | graham dot king at scu dot edu dot au |
| Editors | Harsa, Hayriye S. |
| Year published | 2022 |
| Volume and issue | 9 |
| Article # | 928837 |
| DOI | 10.3389/fnut.2022.928837 |
| ISSN | 2296-861X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fnut.2022.928837/full |
| Download | https://www.frontiersin.org/articles/10.3389/fnut.2022.928837/pdf (PDF) |
Abstract
Informed policy and decision-making for food systems, nutritional security, and global health would benefit from standardization and comparison of food composition data, spanning production to consumption. To address this challenge, we present a formal controlled vocabulary of terms, definitions, and relationships within the Compositional Dietary Nutrition Ontology (CDNO) that enables description of nutritional attributes for material entities contributing to the human diet. We demonstrate how ongoing community development of CDNO classes can harmonize trans-disciplinary approaches for describing nutritional components from food production to diet.
Keywords: dietary composition, food composition, ontologies, nutritional security, FAIR data, knowledge representation, human health
Introduction
Food production and supply systems affect human nutrition and health in personalized and global contexts.[1] However, nutrition-based decisions and data are seldom integrated along the production and supply chain. This information may affect selection of cultivars and conservation of genetic resources, the management of food supply, processing and distribution, and analysis of dietary consumption patterns segmented by various demographics.[2] Although various conventions exist for naming individual chemicals and physical attributes of dietary components, comparison of data and feedback within food systems is often constrained by divergence in formal definitions and classifications.[3] The exchange of knowledge and operational data between domains would benefit from a consistent framework that defines nutritional and phytochemical composition, as well as other attributes of food, including their dietary role and physiological function.
Knowledge representation underpins communication, and it is particularly important for sharing complex data and information within and between diverse domains such as crop biodiversity, food supply, and nutrition.[4] Defining and classifying commonly understood terminology facilitates data acquisition, exchange, and interoperability, where formal systems of domain-specific controlled vocabularies such as ontologies contribute to the representation and sharing of complex knowledge.[5] They do this by defining terms with human-readable definitions alongside machine-readable relationships that facilitate the annotation, exchange, analysis, and interpretation of data.[6] Establishment of clearly defined ontology classes representing domain-specific terminology is the first step to building common platforms that are of practical value to data curators and to end-users searching for relevant information. An approachable lexical representation of objects or concepts from different perspectives, which also helps reduce ambiguities in terminology for non-specialists, is particularly important for describing datasets in food supply chains[7] (Supplementary Figure 1). For instance, nutritional composition may vary depending on factors such as cultivars, cultivation systems, processing variables, food storage, and preparation. Moreover, there is a need to distinguish between individual chemical components and the method by which their concentration is determined. In many standard food composition tables (FCTs) and databases (FCDBs), such information is often conflated or absent.[8]
The Open Biomedical and Biological Ontologies Foundry and Library (OBO) is responsible for the establishment and development of a wide range of formal vocabularies in the life sciences and related domains.[9] The OBO includes the ontology for Chemical Entities of Biological Interest (ChEBI)[10], which provides a valuable resource for structured sets of chemical definitions. OBO principles emphasize the value of reusing terms (formally known as "classes" or "properties") between ontologies. From these seeds the Compositional Dietary Nutrition Ontology (CDNO)[4] was born.
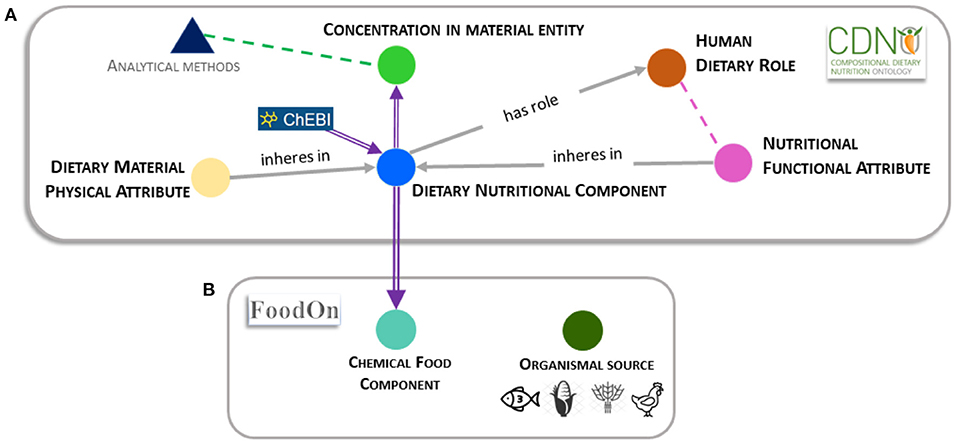
The development of the CDNO[4] was prompted by the need to follow the FAIR principles (findable, accessible, interoperable, and reusable)[11] of data sharing. CDNO was initially focused on vocabulary to describe nutritional components in plant-derived materials contributing to human diet, and particularly those that may vary according to crop variety or within genetic resource collections.[2][4] However, we found that the structured reusable definitions of nutritional components were equally applicable to a wide range of food raw materials derived from livestock, fish, or any other organic or inorganic source described in the Food Ontology (FoodOn)[12] (Figure 1B).
|
Methods
While developing and expanding CDNO, we have followed the OBO principles[13], which emphasize community development of interoperable ontologies. We focused on reuse and import of existing OBO terms, as well as ensuring open discussion within the CDNO GitHub repository.[14] In order to generate terms that are subclasses of CDNO "dietary nutritional component" [CDNO:0200001], a modified version of the Crop Dietary Nutrition Data Framework (CDN-DF) v.1.0 by Halimi et al.[15] was used, with definition and organization of additional terms arising from discussions with plant chemist domain specialists and curators from the International Network of Food Data Systems (INFOODs) collated by the Food and Agriculture Organization (FAO)[16] , U.S. Department of Agriculture (USDA) FoodData Central[17], and the European Food Information Resource (EuroFIR)[18] food composition databases and repositories. The CDN-DF v.2.0 was used as an input for a Python script that parsed the CDN-DF_v.2.0.xlsx into the nutritional_components_framework.csv and sugar_derivatives.csv files, which were converted into input files for ROBOT templates. These templates were used to generate a revised organization of classes/terms compiled into the reference CDNO in a Web Ontology Language (OWL)[19] file. Dietary nutritional components not present in the ChEBI were proposed and accepted as new entities using the ChEBI submission tool and imported into CDNO. The remaining terms that did not fit within the ChEBI scope were formally defined in CDNO, supported by reference to peer reviewed literature and authoritative online resources. These terms were described by following existing ontology definition guidelines for development of genus-differentia definitions.[20] The class "concentration of dietary nutritional component in material entity" [CDNO:0200001], as well as its subclasses, were created using a Dead Simple OWL Design Pattern (DOS-DP)[21] modified from The Environment Ontology (ENVO).[22][23] The DOS-DP combined terms from the Phenotype and Trait Ontology (PATO)[24], CDNO, and the Basic Formal Ontology (BFO)[25] with OWL equivalence axioms. The remaining major classes were proposed and discussed via the CDNO GitHub issue forum[14] and in online workshops and seminars.
The CDNO ontology and accompanying code was initially created using the Ontology-Development-Kit (ODK)[26], and later versions of CDNO were developed using the templates module from the ROBOT software.[27] The reference CDNO OWL file and the source code are available from the Github CDNO repository.[14] Additional database tables were added to the core CropStoreDB MySQL schema[28] to manage different nutritional data sources, along with an "ontology register" lookup table to CDNO, FoodOn, ChEBI, NCBI taxon[29] and Plant Ontology (PO)[30] terms.
Results and discussion
CDNO is registered as part of the OBO Foundry, with terms and definitions searchable via Ontobee[31] or the Ontology Lookup Service (OLS).[32] Version 2.2 of the CDNO comprises five top level classes: "dietary nutritional component" [CDNO:0000001], "concentration of dietary nutritional component in material entity" [CDNO:0200001], "nutritional functional attribute" [CDNO:0300001], "dietary material physical attribute" [CDNO:0400001], and "human dietary role" [CDNO:0500001] (Figure 1A, above).
While dietary nutrients within food substrates are often present as complex and dynamic physical and chemical structures or mixtures, food labelling and FCTs/FCDBs typically represent proximate and individual chemical components, alongside properties such as energy. Within CDNO, the primary "dietary nutritional component" [CDNO:0000001] class is formally defined as, "A material entity taken in by an organism that contributes to the survival, growth, development, or other biological function of itself, its bionts, or its holobionts." This class is structured with 10 subclasses corresponding to the major commonly used proximate classifications of chemical food composition, such as proteins, carbohydrates, and vitamins. The component terms are organized in an acyclic hierarchical structure, with up to five additional levels. At present (v3.1), this class includes over 685 chemical terms, including 29 unique CDNO nutritional chemical components and their definitions. Within FoodOn, the "dietary nutritional component" [CDNO:0000001] class structure has been imported as a subclass of "chemical food component" [FOODON:03411041] class (Figure 1B). A similar hierarchical classification of dietary nutritional components that lacked ontological relationships and definitions had previously been proposed by the EuroFIR project.[18][33] The current versions of EuroFIR thesauri are available online.[34] This was shared following exchange of the original CDNO framework.
Ensuring interoperability of terms defined within CDNO, along with their labels and synonyms, requires ongoing consultation with a range of specialists from different domains. The "dietary nutritional component" [CDNO:0200001] class imports many terms defined within ChEBI, where relationships are primarily determined by formal chemical classifications. However, we were keen to establish a hierarchy that focuses on and accommodates terms organized according to sub-categories recognized by nutritionists and different domain experts, such as food scientists and chemists. We generated and defined subclasses as required, and we included synonyms used in different English-speaking countries. As an example, the term "available carbohydrate" [CDNO:0000003] has the synonym "digestible carbohydrate" according to Englyst et al.[35], but they should not be confused with the term "total carbohydrate" used in some food tables. The latter term is used in the USDA FCT to refer to a specific method used for carbohydrate determination, calculated by subtraction of the sum of the crude protein, total fat, moisture, and ash from the total weight of the food.[36] In order to accommodate such conceptual discrepancies and reduce ambiguity, the term "concentration of carbohydrate in material entity" [CDNO:0200005] can be used to refer to total carbohydrate, without making any assumption as to a specific type of carbohydrates.
Context and use of major classes
A major intended use of the "dietary nutritional component" [CDNO:0000001] class is to harmonize the annotation and exchange of dietary composition datasets from a diverse range of sources that quantify concentration of chemical nutritional components[37] (Figure 1; Supplementary Figure 2). These may include data generated by analytical laboratories for production, reference, and research (Figure 1A, above), as well as derived from existing FCTs/FCDBs or food labeling. Such data may also be used when evaluating evidence in relation to dietary role. The "concentration of dietary nutritional component in material entity" [CDNO:0200001] class is formally defined as, "The concentration of dietary nutritional component when measured in some material entity." In addition, the "dietary material physical attribute" [CDNO:0400001] class is defined as, "A physical property that inheres in a food material or one or more dietary nutritional components." This enables a formal distinction to be made between chemical components and physical properties (or qualities) such as "potential energy" that may inhere in a food material. At present, in most FCTs/FCDBs the tag for “energy” appears equivalent to or alongside chemical components such as sugars (Figure 1A, above).
We make an important distinction between the class "concentration of dietary nutritional component in material entity" [CDNO:0200001] and terms used to describe the analytical method, by which a specific concentration is established. Diverse methods and units of measurement are associated with quantitative data in research literature, for supply chain quality assurance and control, or to inform FCTs and labeling (Figure 1B, above). This requires appropriate vocabulary (potentially in an independent "analytical methods" class) to describe the distinct steps in the process to quantify concentration (Figure 1A, above), including methodologies and protocols used for sampling, extraction, and analysis that may re-use terms from existing ontologies such as the Chemical Method Ontology (CHMO)[38] and the Ontology for Biomedical Investigations (OBI).[39] Many (FCT/FCDB) tags conflate methods used and nutritional components measured, such as INFOODs tag names [GLYCERA], defined as "glycerides, total; determined by analysis," [LACSM] as "lactose; expressed in monosaccharide equivalents," or [CHOT] as "carbohydrate, total; calculated by summation." In the future, such tags could be annotated with a combination of a nutritional component and analytical method term IDs.
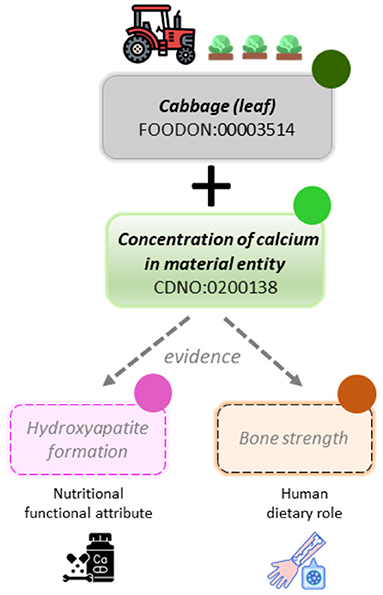
The CDNO team has recently received positive feedback for the classification and description of additional terms to address the increasing clinical, consumer, and market interest in the relationship among the nutritional composition of food and ingredients, their provenance, and their ability to affect personal and public health outcomes. There is a growing but dispersed evidence base associated with "functional foods" and "nutraceuticals." We therefore sought to provide a clear framework that distinguishes among chemical components and physical properties of material entities, the functional attributes they may possess, and any proposed associated human dietary role (Figure 1B, above, and Figure 2). The CDNO "nutritional functional attribute" [CDNO:0300001] class hierarchy (Figure 1A, above) is therefore defined as, "A functional attribute that inheres in one or more dietary nutritional components (or food material) and may contribute to a dietary role." This provides a structured vocabulary that allows description of quantifiable knowledge (Figure 1A, above), with terms such as "antioxidant status" or "glycemic index."
|
In order to represent the distinct concepts relating to potential role in the context of health and wellbeing, we then defined the "human dietary role" [CDNO:0500001] class as, "A biological role that may be assigned to a dietary nutritional component based on evidence, supported at the levels of molecular interaction, cellular process, or physiological role." The value of establishing these distinct classes is demonstrated by the ambiguity associated with use of the word "vitamin," which may refer either to a role [CHEBI:33229] and/or to a chemical entity [CDNO:0000014], depending on context. It is recognized that any conjecture made in relation to role [BFO:0000023] is dependent on an evidence base (Figure 2B, above), and so the terms defined within the "human dietary role" class hierarchy are made available primarily for data curators and specialists to associate or annotate with evidence-based datasets. Moreover, a role may be dependent on many variables, including but not limited to concentration (dose), physical form (bioavailability), demographic, genetic, developmental stage, and/or health status of the human subject, as well as intake of other dietary components. Such variables may be defined within other OBO ontologies such as the Ontology for Nutritional Studies (ONS)[43], which provides a framework for evidence-based studies structured according to various parameters, or the Environmental Conditions, Treatments, and Exposures Ontology (ECTO), which supports modelling of exposure processes such as dietary exposures.[44] We anticipate that further development of the CDNO classes described above may benefit from reuse of terms from additional OBO ontologies such as the Experimental Factor Ontology (EFO)[40], the Human Phenotype Ontology (HP)[41], OBI, and the Ontology of Biological Attributes (OBA).[42] However, substantial work between domain experts and OBO ontology curators will be required to resolve any discrepancies and allocate appropriate terms to the "nutritional functional attribute" or "human dietary role" classes (Figure 2, above).
CDNO for data curation and retrieval
The CDNO is a live open-source project that encourages regular discussion and enhancements to be proposed by stakeholders in the food supply and nutritional domain. Current updates by the CDNO developers are made in consultation with the OBO community. The value of a common vocabulary is demonstrated by the ease with which specific terms may be associated with distinct data sources. Since CDNO is expected to facilitate the compilation and analysis of a diverse range of datasets, we present a use case interface that demonstrates how variation in the concentration of nutritional components may be compared between datasets from different sources and levels of abstraction (Supplementary Figure 2).
Analytical samples may be derived from any stage in the characterization of biodiversity, plant breeding, cultivar improvement, and deployment, through the food production, processing and supply chains, as well as from food storage, preparation, consumption, and digestion (Supplementary Figure 1; Figure 1A, above). In many cases, the "concentration of dietary nutritional component in material entity" [CDNO:0200001] terms would be used to annotate data in a curation pipeline in conjunction with FoodOn terms that define organismal source (e.g., from crops, livestock, fisheries) and organismal part (e.g., grain, liver, fin) (Figure 1B, above).
In order to demonstrate a use case, we curated data that used a series of metadata terms to describe specific sets of nutritional component concentration data. This included the FoodOn "Food product by organism" [FOODON:00002381] class (Figure 1, above), and the National Center for Biotechnology Information (NCBI) organismal classification ontology (NCBITaxon) entity (Supplementary Figure 2). We then developed an online data retrieval interface that allows selection of specific CDNO terms to filter and access multiple sources of nutritional data derived from a crop biodiversity database[45], a national food composition database[46], and a geospatial dietary nutrition study[47] (Supplementary Figure 2).
One valuable outcome of generating an ontology is the opportunity to include structured annotation within the reference OWL file. This may include authoritative citations, as well as formal cross-references to published databases and other reference data sources. We therefore incorporated provisional cross-references between specific terms and widely used FCT/FCDB tagnames/code numbers. For example, the term "concentration of proline in material entity" [CDNO:0200062] was associated with the tagname "PRO" from INFOODs and the code "1226" from the USDA. Ongoing maintenance of this feature will facilitate the harmonization of vocabularies used in different FCDBs.
Conclusion
CDNO is a new open-source, community-developed, web-accessible vocabulary providing a formal representation of commonly used dietary and nutritional terminology. Ongoing development of this ontology[48] will contribute toward data sharing and interoperability, particularly for initiatives where a wide range of foodstuffs are analyzed to diversify diet and agricultural production.[49] We anticipate that extending the range and harmonization of terminologies in food systems will facilitate the sourcing and management of nutritional resources and stimulate development of information-led markets.[50][51] In particular, there is scope for large-scale data integration that enables downstream meta-analyses to complement advances in human, crop, and livestock genomics and high-throughput analytical chemistry.
Supplementary information
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.928837/full#supplementary-material
Supplementary Figure 1. Representation of nutritional information in the continuum between agriculture and health. Knowledge is represented in the context of different domains that may contribute to variation in nutritional outcomes. The symbols under each domain correspond to each of the major CDNO and FoodOn classes represented in Figure 1 (above).
Supplementary Figure 2. Use case of CDNO terms used to mine and compare data from diverse sources. Online graphical interface developed to demonstrate how the "concentration of dietary nutritional component in material entity" [CDNO:0200001] classes are associated with quantitative data derived from (i) genetic resources, (ii) food composition tables, and (iii) a geo-spatial study. Data associated with distinct data sources (see below) have been curated within a relational schema to enable semantic search and filtering based on annotation of key records, where ontology terms are managed within an "ontology register" table. The user may navigate the hierarchical class tree (here, CDNO v3.0) and select (green solid circle) a nutritional component (e.g., CDNO:0200138 - "calcium concentration"). Food or crop groups may be selected (olive solid circle) and then refined to, for example, specific crops or organismal parts used for food. Data from the filtered datasets may be represented by a combination of box-whisker plots, frequency distributions, or single values. Calcium concentration data were sourced to represent variation within (i) a crop biodiversity collection representing the vegetable Brassica oleracea genepool[52], the Brassica Information Portal[45]; (ii) vegetables and vegetable products from a food composition database, the Canadian Nutrient File[46][53]; and (iii) a geospatial study where crop-based edible food items were sampled from multiple locations in Malawi, with data presented for variation in the underutilized crop Moringa oleifera.[47] Recorded values have been adjusted to enable direct comparison on a consistent y-axis, as the original units varied according to each study, as (i) %, (ii) mg/100g, and (iii) mg per kg. For (ii), as for many food composition databases, only single values per nutrient component per food are available[15], although the number of original samples from which the mean value is derived is stated.
Acknowledgements
The authors thank Edward Joy for valuable discussion about the geospatial nutritional data. The authors also thank the Open Biomedical Ontologies personnel who participated in the development, release, and constant improvement of this work. The authors also thank Naomi Fukagawa from the USDA for providing feedback on the initial CDN-DF framework.
Author contributions
LA-H, KB, RW, DD, RM, and GK were members of the working group that conceptualized and design of the CDNO. LA-H, KB, RW, DD, RM, ML, MW, LC, AMa, AMø, JI, LS, XZ, BB-F, PM, SF, AS, LL, and GK provided significant analysis and interpretation of research data. LA-H and GK wrote the first draft of the manuscript. KB and LA-H created the codebase for the CDNO. All authors provided critical revisions on manuscript drafts and approved the final manuscript.
Funding
LA-H performed this work as part of a scholarship from Southern Cross University and Crops For the Future, with additional and subsequent post-doc funding from WISEcode, LLC.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://github.com/Southern-Cross-Plant-Science/cdno.
Conflict of interest
PM and AS are employed by WISECode, LLC. This study received funding from WISECode, LLC. The funder had the following involvement with the study: provision of domain-specific know-how in establishing scope of the ontology and classes, along with key insights into practical end use in data management. SF was employed by Nutrient Institute, LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- ↑ Fanzo, Jessica; Bellows, Alexandra L; Spiker, Marie L; Thorne-Lyman, Andrew L; Bloem, Martin W (4 January 2021). "The importance of food systems and the environment for nutrition" (in en). The American Journal of Clinical Nutrition 113 (1): 7–16. doi:10.1093/ajcn/nqaa313. ISSN 0002-9165. PMC PMC7717136. PMID 33236086. https://academic.oup.com/ajcn/article/113/1/7/6000654.
- ↑ 2.0 2.1 Azman Halimi, Razlin; Barkla, Bronwyn J.; Mayes, Sean; King, Graham J. (1 April 2019). "The potential of the underutilized pulse bambara groundnut (Vigna subterranea (L.) Verdc.) for nutritional food security" (in en). Journal of Food Composition and Analysis 77: 47–59. doi:10.1016/j.jfca.2018.12.008. https://linkinghub.elsevier.com/retrieve/pii/S088915751830766X.
- ↑ Chan, Lauren; Vasilevsky, Nicole; Thessen, Anne; McMurry, Julie; Haendel, Melissa (25 January 2021). "The landscape of nutri-informatics: a review of current resources and challenges for integrative nutrition research" (in en). Database 2021: baab003. doi:10.1093/database/baab003. ISSN 1758-0463. PMC PMC7833928. PMID 33494105. https://academic.oup.com/database/article/doi/10.1093/database/baab003/6119904.
- ↑ 4.0 4.1 4.2 4.3 Andrés‐Hernández, Liliana; Baten, Abdul; Azman Halimi, Razlin; Walls, Ramona; King, Graham J. (1 March 2020). "Knowledge representation and data sharing to unlock crop variation for nutritional food security" (in en). Crop Science 60 (2): 516–529. doi:10.1002/csc2.20092. ISSN 0011-183X. https://onlinelibrary.wiley.com/doi/10.1002/csc2.20092.
- ↑ Munir, Kamran; Sheraz Anjum, M. (1 July 2018). "The use of ontologies for effective knowledge modelling and information retrieval" (in en). Applied Computing and Informatics 14 (2): 116–126. doi:10.1016/j.aci.2017.07.003. https://linkinghub.elsevier.com/retrieve/pii/S2210832717300649.
- ↑ Gan, Mingxin; Dou, Xue; Jiang, Rui (2013). "From Ontology to Semantic Similarity: Calculation of Ontology-Based Semantic Similarity" (in en). The Scientific World Journal 2013: 1–11. doi:10.1155/2013/793091. ISSN 1537-744X. PMC PMC3603583. PMID 23533360. http://www.hindawi.com/journals/tswj/2013/793091/.
- ↑ Davis, Kyle Frankel; Downs, Shauna; Gephart, Jessica A. (1 January 2021). "Towards food supply chain resilience to environmental shocks" (in en). Nature Food 2 (1): 54–65. doi:10.1038/s43016-020-00196-3. ISSN 2662-1355. http://www.nature.com/articles/s43016-020-00196-3.
- ↑ Greenfield, Heather; Southgate, D. A. T. (2003). Food composition data: production, management, and use. Food and Agriculture Organization of the United Nations. Rome: FAO. ISBN 978-92-5-104949-5. OCLC ocm54831428. https://www.worldcat.org/title/mediawiki/oclc/ocm54831428.
- ↑ Jackson, Rebecca; Matentzoglu, Nicolas; Overton, James A; Vita, Randi; Balhoff, James P; Buttigieg, Pier Luigi; Carbon, Seth; Courtot, Melanie et al. (26 October 2021). "OBO Foundry in 2021: operationalizing open data principles to evaluate ontologies" (in en). Database 2021: baab069. doi:10.1093/database/baab069. ISSN 1758-0463. PMC PMC8546234. PMID 34697637. https://academic.oup.com/database/article/doi/10.1093/database/baab069/6410158.
- ↑ Degtyarenko, K.; de Matos, P.; Ennis, M.; Hastings, J.; Zbinden, M.; McNaught, A.; Alcantara, R.; Darsow, M. et al. (23 December 2007). "ChEBI: a database and ontology for chemical entities of biological interest" (in en). Nucleic Acids Research 36 (Database): D344–D350. doi:10.1093/nar/gkm791. ISSN 0305-1048. PMC PMC2238832. PMID 17932057. https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkm791.
- ↑ Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, I. Jsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship". Scientific Data 3: 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. PMC 4792175. PMID 26978244. https://pubmed.ncbi.nlm.nih.gov/26978244.
- ↑ Dooley, Damion M.; Griffiths, Emma J.; Gosal, Gurinder S.; Buttigieg, Pier L.; Hoehndorf, Robert; Lange, Matthew C.; Schriml, Lynn M.; Brinkman, Fiona S. L. et al. (1 December 2018). "FoodOn: a harmonized food ontology to increase global food traceability, quality control and data integration" (in en). npj Science of Food 2 (1): 23. doi:10.1038/s41538-018-0032-6. ISSN 2396-8370. PMC PMC6550238. PMID 31304272. http://www.nature.com/articles/s41538-018-0032-6.
- ↑ OBO Technical WG. "Principles: Overview". OBO Foundry. OBO Technical WG. https://obofoundry.org/principles/fp-000-summary.html. Retrieved 20 January 2022.
- ↑ 14.0 14.1 14.2 Andrés-Hernández, L. (2022). "Southern-Cross-Plant-Science / cdno". GitHub. https://github.com/Southern-Cross-Plant-Science/cdno. Retrieved 20 January 2022.
- ↑ 15.0 15.1 Azman Halimi, Razlin; Barkla, Bronwyn J; Andrés‐Hernandéz, Liliana; Mayes, Sean; King, Graham J (15 March 2020). "Bridging the food security gap: an information‐led approach to connect dietary nutrition, food composition and crop production" (in en). Journal of the Science of Food and Agriculture 100 (4): 1495–1504. doi:10.1002/jsfa.10157. ISSN 0022-5142. https://onlinelibrary.wiley.com/doi/10.1002/jsfa.10157.
- ↑ Charrondiere, U. Ruth; Burlingame, Barbara (1 June 2011). "Report on the FAO/INFOODS Compilation Tool: A simple system to manage food composition data" (in en). Journal of Food Composition and Analysis 24 (4-5): 711–715. doi:10.1016/j.jfca.2010.09.006. https://linkinghub.elsevier.com/retrieve/pii/S0889157510002620.
- ↑ Haytowitz, David B.; Pehrsson, Pamela R. (1 January 2018). "USDA’s National Food and Nutrient Analysis Program (NFNAP) produces high-quality data for USDA food composition databases: Two decades of collaboration" (in en). Food Chemistry 238: 134–138. doi:10.1016/j.foodchem.2016.11.082. https://linkinghub.elsevier.com/retrieve/pii/S0308814616319331.
- ↑ 18.0 18.1 Moller, A; Unwin, I; Becker, W; Ireland, J (1 August 2007). "EuroFIR's food databank systems for nutrients and bioactives" (in en). Trends in Food Science & Technology 18 (8): 428–433. doi:10.1016/j.tifs.2007.02.003. https://linkinghub.elsevier.com/retrieve/pii/S0924224407000490.
- ↑ Bechhofer, Sean (2009), Liu, Ling; Özsu, M. Tamer, eds., "OWL: Web Ontology Language" (in en), Encyclopedia of Database Systems (Boston, MA: Springer US), doi:10.1007/978-0-387-39940-9_1073, ISBN 978-0-387-35544-3, http://link.springer.com/10.1007/978-0-387-39940-9_1073
- ↑ Seppälä, S.; Ruttenberg, A.; Smith, B. (2017). "Guidelines for writing definitions in ontologies". Ciência da Informação 46 (1): 73–88. doi:10.18225/ci.inf..v46i1.4015. https://revista.ibict.br/ciinf/article/view/4015.
- ↑ Osumi-Sutherland, David; Courtot, Melanie; Balhoff, James P.; Mungall, Christopher (1 December 2017). "Dead simple OWL design patterns" (in en). Journal of Biomedical Semantics 8 (1): 18. doi:10.1186/s13326-017-0126-0. ISSN 2041-1480. PMC PMC5460348. PMID 28583177. https://jbiomedsem.biomedcentral.com/articles/10.1186/s13326-017-0126-0.
- ↑ Buttigieg, Pier; Morrison, Norman; Smith, Barry; Mungall, Christopher J; Lewis, Suzanna E; the ENVO Consortium (2013). "The environment ontology: contextualising biological and biomedical entities" (in en). Journal of Biomedical Semantics 4 (1): 43. doi:10.1186/2041-1480-4-43. ISSN 2041-1480. PMC PMC3904460. PMID 24330602. http://jbiomedsem.biomedcentral.com/articles/10.1186/2041-1480-4-43.
- ↑ Buttigieg, Pier Luigi; Pafilis, Evangelos; Lewis, Suzanna E.; Schildhauer, Mark P.; Walls, Ramona L.; Mungall, Christopher J. (1 December 2016). "The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation" (in en). Journal of Biomedical Semantics 7 (1): 57. doi:10.1186/s13326-016-0097-6. ISSN 2041-1480. PMC PMC5035502. PMID 27664130. https://jbiomedsem.biomedcentral.com/articles/10.1186/s13326-016-0097-6.
- ↑ Gkoutos, Georgios V; Green, Eain CJ; Mallon, Ann-Marie; Hancock, John M; Davidson, Duncan (2004). "[No title found"]. Genome Biology 6 (1): R8. doi:10.1186/gb-2004-6-1-r8. PMC PMC549069. PMID 15642100. http://genomebiology.biomedcentral.com/articles/10.1186/gb-2004-6-1-r8.
- ↑ Smith, B. (2012). "On Classifying Material Entities in Basic Formal Ontology". Interdisciplinary Ontology: Proceedings of the Third Interdisciplinary Ontology Meeting: 1–13. https://philarchive.org/rec/SMIOCM.
- ↑ OBO Technical WG (2012). "Creating a new Ontology using the ODK". OBO Foundry. https://ontology-development-kit.readthedocs.io/en/latest/NewRepo.html. Retrieved 12 June 2020.
- ↑ Jackson, Rebecca C.; Balhoff, James P.; Douglass, Eric; Harris, Nomi L.; Mungall, Christopher J.; Overton, James A. (1 December 2019). "ROBOT: A Tool for Automating Ontology Workflows" (in en). BMC Bioinformatics 20 (1): 407. doi:10.1186/s12859-019-3002-3. ISSN 1471-2105. PMC PMC6664714. PMID 31357927. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-019-3002-3.
- ↑ Leibovici, Didier G.; Anand, Suchith; Santos, Roberto; Mayes, Sean; Ray, Rumiana; Al-Azri, Masoud; Baten, Abdul; King, Graham et al. (1 December 2017). "Geospatial binding for transdisciplinary research in crop science: the GRASPgfs initiative" (in en). Open Geospatial Data, Software and Standards 2 (1): 20. doi:10.1186/s40965-017-0034-3. ISSN 2363-7501. http://opengeospatialdata.springeropen.com/articles/10.1186/s40965-017-0034-3.
- ↑ Schulz, S.; Stenzhorn, H.; Boeker, M. (1 July 2008). "The ontology of biological taxa" (in en). Bioinformatics 24 (13): i313–i321. doi:10.1093/bioinformatics/btn158. ISSN 1367-4803. PMC PMC2718636. PMID 18586729. https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btn158.
- ↑ Jaiswal, Pankaj; Ware, Doreen; Ni, Junjian; Chang, Kuan; Zhao, Wei; Schmidt, Steven; Pan, Xiaokang; Clark, Kenneth et al. (2002). "Gramene: Development and Integration of Trait and Gene Ontologies for Rice" (in en). Comparative and Functional Genomics 3 (2): 132–136. doi:10.1002/cfg.156. ISSN 1531-6912. PMC PMC2447246. PMID 18628886. http://www.hindawi.com/journals/ijg/2002/432674/abs/.
- ↑ "Compositional Dietary Nutrition Ontology". Ontobee. He Group. https://ontobee.org/ontology/CDNO. Retrieved 20 January 2022.
- ↑ "Compositional Dietary Nutrition Ontology". Ontology Lookup Service. EMBL-EBI. https://www.ebi.ac.uk/ols/ontologies/cdno. Retrieved 20 March 2022.
- ↑ Møller, A.; Unwin, I.D.; Ireland, J. et al. (2008). "The EuroFIR Thesauri 2008" (PDF). ISBN 9788792125095. http://archives.foodcomp.info/eurofir/Downloads/Thesauri/EuroFIR%20Thesauri%202008.pdf.
- ↑ European Food Information Resource (2022). "EuroFIR Thesauri". European Food Information Resource. https://www.eurofir.org/our-resources/eurofir-thesauri/. Retrieved 20 January 2022.
- ↑ Englyst, K N; Liu, S; Englyst, H N (1 December 2007). "Nutritional characterization and measurement of dietary carbohydrates" (in en). European Journal of Clinical Nutrition 61 (S1): S19–S39. doi:10.1038/sj.ejcn.1602937. ISSN 0954-3007. https://www.nature.com/articles/1602937.
- ↑ Cummings, J H; Stephen, A M (1 December 2007). "Carbohydrate terminology and classification" (in en). European Journal of Clinical Nutrition 61 (S1): S5–S18. doi:10.1038/sj.ejcn.1602936. ISSN 0954-3007. https://www.nature.com/articles/1602936.
- ↑ Finglas, Paul M.; Berry, Rachel; Astley, Siân (1 September 2014). "Assessing and Improving the Quality of Food Composition Databases for Nutrition and Health Applications in Europe: The Contribution of EuroFIR" (in en). Advances in Nutrition 5 (5): 608S–614S. doi:10.3945/an.113.005470. ISSN 2156-5376. PMC PMC4188244. PMID 25469406. https://academic.oup.com/advances/article/5/5/608S/4565780.
- ↑ Batchelor, C. (2021). "rsc-ontologies / rsc-cmo". GitHub. https://github.com/rsc-ontologies/rsc-cmo. Retrieved 13 February 2021.
- ↑ Bandrowski, Anita; Brinkman, Ryan; Brochhausen, Mathias; Brush, Matthew H.; Bug, Bill; Chibucos, Marcus C.; Clancy, Kevin; Courtot, Mélanie et al. (29 April 2016). Xue, Yu. ed. "The Ontology for Biomedical Investigations" (in en). PLOS ONE 11 (4): e0154556. doi:10.1371/journal.pone.0154556. ISSN 1932-6203. PMC PMC4851331. PMID 27128319. https://dx.plos.org/10.1371/journal.pone.0154556.
- ↑ 40.0 40.1 Malone, James; Adamusiak, Tomasz; Holloway, Ele; Parkinson, Helen (25 September 2009). "Developing an application ontology for annotation of experimental variables – Experimental Factor Ontology" (in en). Nature Precedings 10. doi:10.1038/npre.2009.3806.1. ISSN 1756-0357. http://www.nature.com/articles/npre.2009.3806.1.
- ↑ 41.0 41.1 Robinson, Peter N.; Köhler, Sebastian; Bauer, Sebastian; Seelow, Dominik; Horn, Denise; Mundlos, Stefan (1 November 2008). "The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease" (in en). The American Journal of Human Genetics 83 (5): 610–615. doi:10.1016/j.ajhg.2008.09.017. PMC PMC2668030. PMID 18950739. https://linkinghub.elsevier.com/retrieve/pii/S0002929708005351.
- ↑ 42.0 42.1 "TermGenie for the ontology of biological attributes (OBA)". 2016. http://oba.termgenie.org/. Retrieved 09 March 2022.
- ↑ the ENPADASI consortium; Vitali, Francesco; Lombardo, Rosario; Rivero, Damariz; Mattivi, Fulvio; Franceschi, Pietro; Bordoni, Alessandra; Trimigno, Alessia et al. (1 December 2018). "ONS: an ontology for a standardized description of interventions and observational studies in nutrition" (in en). Genes & Nutrition 13 (1): 12. doi:10.1186/s12263-018-0601-y. ISSN 1555-8932. PMC PMC5928560. PMID 29736190. https://genesandnutrition.biomedcentral.com/articles/10.1186/s12263-018-0601-y.
- ↑ Chan, Lauren; Thessen, Anne; Duncan, William D.; Matentzoglu, Nicolas; Schmitt, Charles; Grondin, Cynthia; Vasilevsky, Nicole; McMurry, Julie et al. (15 March 2022). "The Environmental Conditions, Treatments, and Exposures Ontology (ECTO): Connecting Toxicology and Exposure to Human Health and Beyond" (in en). Zenodo. doi:10.5281/ZENODO.6360645. https://zenodo.org/record/6360645.
- ↑ 45.0 45.1 Eckes, Annemarie H.; Gubała, Tomasz; Nowakowski, Piotr; Szymczyszyn, Tomasz; Wells, Rachel; Irwin, Judith A.; Horro, Carlos; Hancock, John M. et al. (12 April 2017). "Introducing the Brassica Information Portal: Towards integrating genotypic and phenotypic Brassica crop data" (in en). F1000Research 6: 465. doi:10.12688/f1000research.11301.1. ISSN 2046-1402. PMC PMC5428495. PMID 28529710. https://f1000research.com/articles/6-465/v1.
- ↑ 46.0 46.1 Deeks, Josephine; Verreault, Marie-France; Cheung, Winnie (1 December 2017). "Canadian Nutrient File (CNF): Update on Canadian food composition activities" (in en). Journal of Food Composition and Analysis 64: 43–47. doi:10.1016/j.jfca.2017.04.009. https://linkinghub.elsevier.com/retrieve/pii/S088915751730100X.
- ↑ 47.0 47.1 Joy, Edward J.M.; Broadley, Martin R.; Young, Scott D.; Black, Colin R.; Chilimba, Allan D.C.; Ander, E. Louise; Barlow, Thomas S.; Watts, Michael J. (1 February 2015). "Soil type influences crop mineral composition in Malawi" (in en). Science of The Total Environment 505: 587–595. doi:10.1016/j.scitotenv.2014.10.038. https://linkinghub.elsevier.com/retrieve/pii/S0048969714014764.
- ↑ "Compositional Dietary Nutrition Ontology (CDNO)". Southern Cross University. https://www.cdno.info/. Retrieved 08 April 2022.
- ↑ Cifelli, Christopher J (8 December 2021). "Looking beyond traditional nutrients: the role of bioactives and the food matrix on health" (in en). Nutrition Reviews 79 (Supplement_2): 1–3. doi:10.1093/nutrit/nuab100. ISSN 0029-6643. PMC PMC8653941. PMID 34879144. https://academic.oup.com/nutritionreviews/article/79/Supplement_2/1/6457114.
- ↑ Mazac, Rachel; Tuomisto, Hanna L. (18 March 2020). "The Post-Anthropocene Diet: Navigating Future Diets for Sustainable Food Systems" (in en). Sustainability 12 (6): 2355. doi:10.3390/su12062355. ISSN 2071-1050. https://www.mdpi.com/2071-1050/12/6/2355.
- ↑ Zeb, Akhtar; Soininen, Juha-Pekka; Sozer, Nesli (1 May 2021). "Data harmonisation as a key to enable digitalisation of the food sector: A review" (in en). Food and Bioproducts Processing 127: 360–370. doi:10.1016/j.fbp.2021.02.005. https://linkinghub.elsevier.com/retrieve/pii/S0960308521000237.
- ↑ Broadley, Martin R.; Hammond, John P.; King, Graham J.; Astley, Dave; Bowen, Helen C.; Meacham, Mark C.; Mead, Andrew; Pink, David A.C. et al. (3 April 2008). "Shoot Calcium and Magnesium Concentrations Differ between Subtaxa, Are Highly Heritable, and Associate with Potentially Pleiotropic Loci in Brassica oleracea" (in en). Plant Physiology 146 (4): 1707–1720. doi:10.1104/pp.107.114645. ISSN 1532-2548. PMC PMC2287345. PMID 18281414. https://academic.oup.com/plphys/article/146/4/1707/6107189.
- ↑ "Canadian Nutrient File (CNF), 2015". Government of Canada. 3 June 2016. https://www.canada.ca/en/health-canada/services/food-nutrition/healthy-eating/nutrient-data/canadian-nutrient-file-2015-download-files.html. Retrieved 20 January 2022.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation and updates to spelling and grammar. In some cases important information was missing from the references, and that information was added.