Journal:Baobab Laboratory Information Management System: Development of an open-source laboratory information management system for biobanking
| Full article title | Baobab Laboratory Information Management System: Development of an open-source laboratory information management system for biobanking |
|---|---|
| Journal | Biopreservation and Biobanking |
| Author(s) | Bendou, Hocine; Sizani, Lunga; Reid, Tim; Swanepoel, Carmen; Ademuyiwa, Toluwaleke; Merino-Martinez, Roxana; Meuller, Heimo; Abayomi, Akin; Christoffels, Alan |
| Author affiliation(s) | University of the Western Cape, B3Africa Consortium, Tygerberg Hospital, Stellenbosch University, H3Africa Consortium, Karolinska Institutet, BBMRI-ERIC Common Service IT |
| Primary contact | Email: alan at sanbi dot ac dot za |
| Year published | 2017 |
| Volume and issue | 15 (2) |
| Page(s) | 116-120 |
| DOI | 10.1089/bio.2017.0014 |
| ISSN | 1947-5543 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://online.liebertpub.com/doi/full/10.1089/bio.2017.0014 |
| Download | http://online.liebertpub.com/doi/pdfplus/10.1089/bio.2017.0014 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
A laboratory information management system (LIMS) is central to the informatics infrastructure that underlies biobanking activities. To date, a wide range of commercial and open-source LIMSs are available, and the decision to opt for one LIMS over another is often influenced by the needs of the biobank clients and researchers, as well as available financial resources. The Baobab LIMS was developed by customizing the Bika LIMS software to meet the requirements of biobanking best practices. The need to implement biobank standard operation procedures as well as stimulate the use of standards for biobank data representation motivated the implementation of Baobab LIMS, an open-source LIMS for biobanking. Baobab LIMS comprises modules for biospecimen kit assembly, shipping of biospecimen kits, storage management, analysis requests, reporting, and invoicing. The Baobab LIMS is based on the Plone web-content management framework. All the system requirements for Plone are applicable to Baobab LIMS, including the need for a server with at least 8 GB RAM and 120 GB hard disk space. Baobab LIMS is a client-server-based system, whereby the end user is able to access the system securely through the internet on a standard web browser, thereby eliminating the need for standalone installations on all machines.
Introduction
Human biobanking refers to the collection, processing, and storage of biospecimens and the collection of associated demographic and clinical data for future research use. The extensive collections of biospecimens throughout Africa collected for either specific research, population studies, or part of normal diagnostics workup were not necessarily collected for prospective use by researchers and practitioners. As a result, such collections might not necessarily have followed or adhered to evolving bioethical paradigms and international biobanking best practices.[1][2]
However, the establishment of the concept of centralized biobanks across Africa through initiatives such as H3Africa (http://www.h3africa.org/consortium/projects), the AIDS Cancer Specimen Resource (ACSR; http://oham.cancer.gov/oham_research/programs/specimen_resource), and the B3Africa (http://www.b3africa.org) projects has highlighted the need for establishing and harmonizing national and regional biobank governance frameworks to address a relatively unregulated access to human and other ecological samples of academic interest in Africa. At the same time, these governance frameworks fall in line with rapidly changing biobanking practices driven by modern technology.[3][4] Similarly, a governance framework for IT infrastructure requirements that underlies a biobank does not exist.
According to the biological material tracking recommendations within the ISBER best practices, a computer-based inventory system should be in place to allow for the tracking and annotation of each incoming biospecimen into the biobank.[1] An LIMS is thus central to the informatics infrastructure that underlies biobanking activities. To date, a wide range of commercial and open-source LIMSs are available, and the decision to opt for one LIMS over another is often influenced by the needs of the biobank clients and researchers, as well as available financial resources.
The National Health Laboratory Services (NHLS)—Stellenbosch University Biobank (NSB), a unit associated with the Division of Haematology at the Faculty of Medicine and Health Sciences, was established in 2012 initially through the ACSR project, and subsequently the NIH H3Africa funding initiative, and required options for an LIMS implementation. The only option at the time was to consider a commercial LIMS because of time constraints to meet the growing need for biobanking services in South Africa. However, access to open-source LIMS software allowed us to consider a longer term implementation that would align with our sustainability plans. Bika LIMS[5] and caTissue (now evolved and known as OpenSpecimien[6]) were identified as long-term options based on input from active software developer and user communities.
Bika LIMS, although not specifically made for human biospecimens, is part of the BIKA software ecosystem that includes BIKA Health for healthcare laboratories and Bika Interlab for interlaboratory proficiency testing. Customization of the Bika LIMS software would provide the benefit of inheriting a range of electronic health record functions that are central to establishing a core facility to support personalized medicine research. The recently funded European project, B3Africa, was established to strengthen IT infrastructure and ethical governance frameworks that would bridge biobanking and biomedical research across Europe and Africa. This funding provided the impetus to revisit the biobanking IT infrastructure at the NSB and to accelerate the development of Baobab LIMS, an open-source LIMS for biobanking, as a strategy to provide a harmonized LIMS as an option for Africa.
A functional specification exercise[7] in 2013 within the context of NSB biobanking requirements identified the following key modules as part of the extension to the existing Bika LIMS software, namely biospecimen kit assembly, shipping of biospecimen kits, storage management, analysis requests, reporting, and invoicing.
Implementation
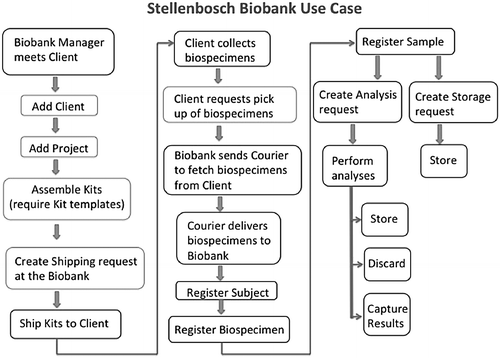
Standard operating procedures (SOPs) associated with biological material inventory management were developed by the NSB team to inform LIMS workflow development. Other SOPs focusing on shipping, labeling, biospecimen procedures, and quality control were also developed in association with other H3Africa biobanks, and they are publicly available (http://h3africa.org/consortium/documents). The collection of SOPs underlies the NSB flowchart of biobank activities (Fig. 1) and subsequently the quality management system. Importantly, even with the use of electronic systems, it is important to keep hard copies of all documentation detailing the biospecimen passage from reception throughout storage to dissemination as a QC check.
|
References
- ↑ 1.0 1.1 International Society for Biological and Environmental Repositories (2012). "2012 Best Practices for Repositories: Collection, Storage, Retrieval, and Distribution of Biological Materials for Research". Biopreservation and Biobanking 10 (2): 79-161. doi:10.1089/bio.2012.1022. PMID 24844904.
- ↑ Abayomi, A.; Christoffels, A.; Grewal, R. et al. (2013). "Challenges of biobanking in South Africa to facilitate indigenous research in an environment burdened with human immunodeficiency virus, tuberculosis, and emerging noncommunicable diseases". Biopreservation and Biobanking 11 (6): 347–354. doi:10.1089/bio.2013.0049. PMID 24835364.
- ↑ Dhai, A. (2013). "Establishing national biobanks in South Africa: The urgent need for an ethico-regulatory framework". The South African Journal of Bioethics & Law 6 (2): 38–39. doi:10.7196/SAJBL.296.
- ↑ de Vries, J.; Abayomi, A.; Brandful, J. et al. (2014). "A perpetual source of DNA or something really different: ethical issues in the creation of cell lines for African genomics research". BMC Medical Ethics 15: 60. doi:10.1186/1472-6939-15-60.
- ↑ "BIKA". Bika Lab Systems (Pty) Ltd. https://www.bikalims.org/. Retrieved 16 August 2016.
- ↑ "OpenSpecimen". Krishagni Solutions Pvt. Ltd. http://www.openspecimen.org/. Retrieved 16 August 2016.
- ↑ NCB-H3A (23 March 2013). "NCB-H3A Cape Town Biobank: Biobank Information Management System - Functional Requirements Overview & Phase I Objectives". South African National Bioinformatics Institute. http://christoffels.sanbi.ac.za/index.php/projects/biobanking. Retrieved 16 August 2016.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.