Journal:An automated dashboard to improve laboratory COVID-19 diagnostics management
| Full article title | An automated dashboard to improve laboratory COVID-19 diagnostics management |
|---|---|
| Journal | Frontiers in Digital Health |
| Author(s) | Maury, Emma; Boldi, Marc-Olivier; Greub, Gilbert; Chavez, Valérie; Jaton, Katia; Opota, Onya |
| Author affiliation(s) | University of Lausanne, Lausanne University Hospital |
| Primary contact | Email: onya dot opota at chuv dot ch |
| Editors | Hochheiser, Harry |
| Year published | 2021 |
| Volume and issue | 3 |
| Article # | 773986 |
| DOI | 10.3389/fdgth.2021.773986 |
| ISSN | 2673-253X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fdgth.2021.773986/full |
| Download | https://www.frontiersin.org/articles/10.3389/fdgth.2021.773986/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Background: In response to the COVID-19 pandemic, our microbial diagnostic laboratory located in a university hospital has implemented several distinct SARS-CoV-2 reverse transcription polymerase chain reaction (RT-PCR) systems in a very short time. More than 148,000 tests have been performed over 12 months, which represents about 405 tests per day, with peaks to more than 1,500 tests per days during the second wave. This was only possible thanks to automation and digitalization, to allow high-throughput, acceptable time to results and to maintain test reliability. An automated dashboard was developed to give access to key performance indicators (KPIs) to improve laboratory operational management.
Methods: RT-PCR data extraction of four respiratory viruses—SARS-CoV-2, influenza A and B, and RSV—\—from our laboratory information system (LIS) was automated. This included age, gender, test result, RT-PCR instrument, sample type, reception time, requester, hospitalization status, etc. Important KPIs were identified and the visualization was achieved using an in-house dashboard based on the R open-source language (R Shiny).
Results: The dashboard is organized into three main parts. The “Filter” page presents all the KPIs, divided into five sections: (i) general and gender-related indicators, (ii) number of tests and positivity rate, (iii) cycle threshold and viral load, (iv) test durations, and (v) not valid results. Filtering allows to select a given period, a dedicated instrument, a given specimen, an age range, or a requester. The “Comparison” page allows custom charting of all the available variables, which represents more than 182 combinations. The “Data” page gives the user access to the raw data in a tabular format, with the possibility of filtering, allowing for a deeper analysis and data download. Information is updated every four hours.
Conclusions: By giving rapid access to a huge number of up-to-date data points, represented using the most relevant visualization types without the burden of timely data extraction and analysis, the dashboard represents a reliable and user-friendly tool for operational laboratory management, improving the decision-making process, resource planning, and quality management.
Keywords: COVID-19, medical microbiology, dashboard, digitalization, operations management, quality management, key performance indicator
Introduction
In December 2019, a new virus causing pneumonia of unknown etiology emerged in China. Its incidence exploded rapidly, first in the Wuhan region (Hubei province), then in the other regions of China and other countries in Southeast Asia. On January 30, 2020, the World Health Organization (WHO) declared this new coronavirus a “public health emergency of international concern.” [1] On the February 20, the first patient was diagnosed in Italy, in the Lombardy region. The epidemic then spread to other European countries, including Switzerland [2], and the first case was admitted to Lausanne University Hospital on February 28. On March 11, 2020 the WHO declared a pandemic, referring to the disease as Coronavirus disease 2019 or COVID-19. [3–5]
To face the COVID-19 pandemic, caused by the virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), diagnostic laboratories had to develop reverse transcription polymerase chain reaction (RT-PCR) tests allowing the detection of SARS-CoV-2 RNA in patients suspected of contracting COVID-19. Our laboratory, the Institute of Microbiology (IMU), located in one of the five teaching hospitals of Switzerland, the Lausanne University Hospital (CHUV), rapidly developed RT-PCR to detect SARS-CoV-2 in clinical specimens. [6] Microbiological diagnosis of SARS-CoV-2 represents one of the pillars of the diagnosis of COVID-19. Indeed, RT-PCR is also the heart of the patient care and epidemic control process and will be the mainstay of several clinical studies.
Although our laboratory has extensive experience in the development of RT-PCR, the introduction of this new parameter represented a challenge in terms of speed of development. [7] It is also the first time that an introduced parameter has been used on such a large scale in such a short time; more than 10,000 tests were carried out in one month in the spring of 2020 [6], and even in a single week during the fall of 2020. This was possible thanks to automation and digitalization, to allow high-throughput and acceptable time to results. [7]. In this context, the IMU set strategies to ensure the quality and reliability of RT-PCR. This included the monitoring of key performance indicators (KPIs) for quality management such as the proportions of positive tests or the virus load, both per day, per instruments, and per requester. These indicators aimed to identify variations not explained by epidemiological changes. Indeed, abnormal variations could be synonymous with pre-analytical problems (e.g., a sampling problem, transport medium, etc.) or even analytical problems (e.g., mutation in the target sequences of PCRs associated with losses of sensitivity or specificities). The IMU also defined KPIs for operations management, such as the turnaround time (TAT). [8]
Before COVID-19, such indicators were monitored periodically, for example in the context of an annual report or retrospective studies. At the beginning of the COVID-19 outbreak, the IMU decided to follow these indicators frequently. Because the manual analyses were time-consuming, the monitoring of analytical and operational KPIs was carried out once a week initially, and then twice a week depending on the period. These analyses were also prone to error, due to multiple sources of information, repeated manual actions (e.g., copy/cut and paste), and the diversity of the data. All this information required a dashboard.
A dashboard is a graphical user interfaces (GUI) with a database. It allows users to retrieve the relevant information—often KPIs—in a specific context by representing the data in a meaningful and usable way. [9] (See Eckerson 2010 [10] for more details.) In management and business contexts, dashboards aim at turning the overwhelming information volume into an informative opportunity [11] and are part of visual analytics, defined by Cook and Thomas as the “science of analytical reasoning facilitated by interactive visual interface.” [12]
Like any other information technology in the healthcare industry, a dashboard is intended to improve efficiency. [13] Dashboards help monitor daily activities [14], such as tracking ongoing operations, a priority in healthcare institutions. [15] Providing easy access to this information helps the team to make better informed decisions [16], which could take a tremendous amount of time without the technology. [17] Correctly designed and built, dashboards improve the institution's efficiency while providing better quality of care. [18] Cheng et al. [19] offer an example of this in their study on how to build a dashboard to track respiratory viruses like influenza.
With COVID-19 spreading fast over the world, the speed at which data are gathered, integrated, and used became central in the management of this crisis by all health-related institution teams. Interactive dashboards appeared to be appropriate to this aim. A famous example remains the one from Johns Hopkins University. [20]. In Switzerland, corona-data.ch [21] is an up-to-date webpage built at the macro level. With the whole healthcare industry being impacted by the pandemic, various topics and workflow areas were analyzed through dashboards, including e-consultations [22], incident command [23], and performance comparisons to similar institutions. [24]
Also, laboratories responsible for testing patients during an outbreak must control some information to ensure the highest quality of results. In particular, it is crucial to define KPIs, for example to better track daily operations (25) and ensure enough testing capacity. Moreover, when the objective is to increase capacity and quality as well as maintaining schedules, providing valuable insights and pieces of information to the laboratory management can be critical. [26]
A dashboard is an expression of a database. Therefore, as mentioned by O'Donnell and David [27], the resulting decision process depends on the information system (IS), the environment, and the user skills. Regarding the dashboard content, there is no consensus on the format of visualizations, with some researchers claiming no effect on the user's judgement [28] or stating a lack of universality in representation [29], or a preference for tabular information [30]. Wilson and Zigurs [31] showed that even the user's preferred format did not necessarily lead to greater performance, except in the case of symbolic and spatial tasks. [32] Choosing the appropriate visualization can be challenging and is subject to various principles. Lengler and Eppler [33] condensed many visualizations into a "Periodic Table of Visualization Methods," with dashboard data displayed according to three categories: boxes, tables, and plots. Each has advantages and drawbacks, the choice being made based on the end user's needs.
In this paper, we present the design, development, and use of a dashboard targeted to a laboratory such as the IMU, located in a teaching hospital and in charge of PCR testing, following the COVID-19 outbreak. This work addresses defining the need for a dashboard, building the dashboard, deploying the tool, and demonstrating its added value in terms of the quality and operations management goals set out by the laboratory. This research also focuses on aspects other than epidemiological matters (patient type, pathogen, period of the year, etc.), which can explain some variation of results in the laboratory. We split these aspects into two main categories: quality issues and management issues.
Materials and methods
RT-PCR and data
RT-PCR for the detection of SARS-CoV-2 from clinical specimens was achieved as previously described using our in-house molecular diagnostic platform (MDx platform), the Cobas SARS-CoV-2 test on the Cobas 6800 instrument (Roche, Basel, Switzerland), and the Xpert Xpress SARS-CoV-2 assay (Cepheid, CA, USA). [6, 34, 35] Viral load was obtained by conversion of the Ct (Cycle threshold) values of the instruments using either a plasmid containing the target sequence of the PCR obtained from RD-Biotech (Besançon, France) or using purified viral RNA, kindly provided by the Institute of Virology of the University of Berlin, la Charite. [34, 35]
The dataset feeding the dashboard is an extract from Molis, a laboratory information system (LIS) used at the IMU. The extract is performed every four hours. The analyses of four respiratory virus of interest were flagged in the system: SARS CoV-2, Influenza A and B, and the respiratory syncytial virus (RSV). A comma-separated values file (csv) with new observations validated in the prior four hours is uploaded in a specific folder to be read through the dashboard.
To date, the system has 148,667 observations of SARS-CoV-2 via RT-PCR. For each specimen (swab, blood, etc.), the available entries include a unique anonymous patient ID, date of birth and gender of the patient, its hospitalization status, an anonymized code of the entity requesting the test (doctors, clinics, other laboratories, etc.), the date-time of the sampling (when available), of the test, and of the result sending as well as the type of sampling (nasopharyngeal or oropharyngeal secretions, blood sample, etc.). The original dataset also contains analysis codes showing the test result (positive, negative, canceled, NOT VALID), the Ct values, the viral quantification (in copy per milliliter, cp/mL), and whether the analysis had to be repeated. Every test is associated with various internal control tests (e.g., extraction, inhibition, etc.). “NOT VALID” results correspond to analyses which do not pass one of the internal quality controls.
A specimen is related to one patient, but a patient may be tested several times. The analysis codes correspond to a test, which is performed for a specific virus, on a specific device (machine used to perform the test), for a targeted gene.
Some cleaning and data wrangling were performed before building the dashboard. Using a matching table shared by the IMU, the analysis codes were renamed using a more user-friendly structure (NOM.VIRUS_TYPE.ANALYSE_APPAREIL_GENE). Then, different measures were extracted, especially on date-time data: the reception duration is the difference between the sampling time and the reception time at the laboratory, the test duration is the difference between the reception time and the results validation time, and the total duration is the sum of the last two. Using date of birth, patients were categorized into age groups with a 10-year window. Similarly, the type of sampling was recoded using wider groups categories, the most present being NPS (nasopharyngeal secretions). Then, each analysis is assigned a “Virus” and a “Device,” corresponding to the non-empty analysis codes described above. Another “Confirmation” variable was added, showing whether the analysis had to be repeated. Finally, four different tables were created, corresponding to each virus present in the dashboard.
Cleaning and wrangling is done automatically every time the dashboard is launched, without any action from the end-user.
Platform
The interactive dashboard was built using R Shiny [36], based on the open-source R programming language v.4.0.2 [37] in the RStudio Interactive Development Environment. [38] CSS and JavaScript were also used to tailor the dashboard to the needs of the end users. The dashboard relies on many R packages:
- shinydashboard 0.7.1 [39] for the page structure;
- plotly 4.9.3 [40] and ggplot2 3.3.3 [41] for interactive graphs;
- DT 0.17 [42] for interactive tables;
- shinyjs 2.0.0 [43] for some custom interaction;
- shinyWidgets 0.5.6 [44] for input objects; and
- readx 1.3.1l [45], plyr 1.8.6 [46], dplyr 1.0.7 [47], lubridate 1.7.9.2 [48], tidyr 1.1.2 [49], stringr 1.4.0 [50], and forcats 0.5.0 [51] for data wrangling.
The dashboard is built in two dimensions (Figure 1). The horizontal dimension is the Target and the vertical dimension the Action. The level of detail and the amount of possibilities increase when the user goes down on the Action sections.
|
As stated above, picking the appropriate visualization can be challenging and is subject to various principles. There are three main components chosen to build the dashboard:
- Infoboxes: This type of visualization gives immediate information on some key metrics. Placed at the top of the dashboard, the user directly sees crucial information. When having several of these, it is important to group and label them appropriately. [52]
- Tables: Columns describe a specific attribute for each row, showing the user a detailed view, ready for a deeper inspection. Filtering and ordering options are available to display the portion of interest of the data.
- Plots: We used both Abela [53] and the data-to-viz.com tool created by Holtz [54], which provide clear tools to select the appropriate chart relative to the data. This dashboard mostly uses Column charts and Stacked Column charts, Scatter plots, Line charts, and Boxplots. The latter has the advantage of displaying many types of information at once, and the end users are accustomed to this format.
Finally, for a quicker adoption and an optimal usage, the dashboard was built in French, the mother tongue of the end users. The dashboard runs locally on every user's computer, in order to prevent from security issues. The data is stored on the secured CHUV's servers, accessible only when authenticated on this network. Its use is restricted to authorized staff members only, whose mission is closely linked to quality management.
Ethics
The data were obtained during a quality enhancement project at our institution. According to national law, the performance of a project and the subsequent publishing of the results can be done without asking the permission of the competent research ethics committee.
Results
Structure and main KPIs of the dashboard
Global structure of the dashboard
The content of the dashboard was guided by three main principles: comprehensive initial briefings, multiple feedback loops, and close monitoring of latest breakthrough discoveries about SARS-CoV-2. At the beginning of the process, the end users—namely the managers of the molecular diagnostic laboratory—formulated their main needs and made suggestions on what to report in the dashboard. This included the main KPIs, such as the number of tests per day as well as some of the inputs that could be entered by the user, for instance filters such as RT-PCR instruments or patients' gender or age. In an Agile-like methodology, applying continuous improvement [55], demos were regularly performed and feedback rapidly implemented. With SARS-CoV-2 being a novel virus, the scientific literature was closely scrutinized to incorporate new relevant elements, such as gender-related indicators, which were included after Ancochea et al. [56] published their research on gender bias in COVID-19 diagnosis.
The current version of the dashboard contains three main pages, detailed in Figure 1: “Filter,” “Comparison,” and “Data.” The Filter page allows the user to select inputs such as date range, gender, age, test result, hospitalization status, device used, confirmation status, type of sample, and type of requesters. On the Comparison page, the user selects variables to appear on graphs and filters the dataset to narrow down comparison subjects. Finally, the Data page also lets the user to filter the dataset to look at individual observations. Overall, the user therefore has a role in filtering observations and in deciding which information is to be represented (Figure 1).
The Filter page
The Filter page presents a total of 10 KPIs and is divided into five sections: general and gender-related indicators, number of tests and positivity rate, viral load, test durations, and “NOT VALID.”
General and gender-related indicators
The 10 KPIs are presented in aggregate form in this section, some of them being detailed in the subsequent sections. The most general figures are shown at the top of the layout and are split into two groups: "General Indicators" and "Gender-related Indicators." The General Indicators grouping has the total number of tests performed for the selected virus, the number of positive tests, the average test duration displayed next to the median duration, the average number of tests per patient, the percentage of patients having to take two or more tests, and the proportion of tests marked as “NOT VALID." The Gender-related Indicators grouping has the sex ratio (positive women out of positive men), the average age, the positivity rate, and the hospitalization rate (Supplementary Figures 1A,B).
Number of tests and positivity rate
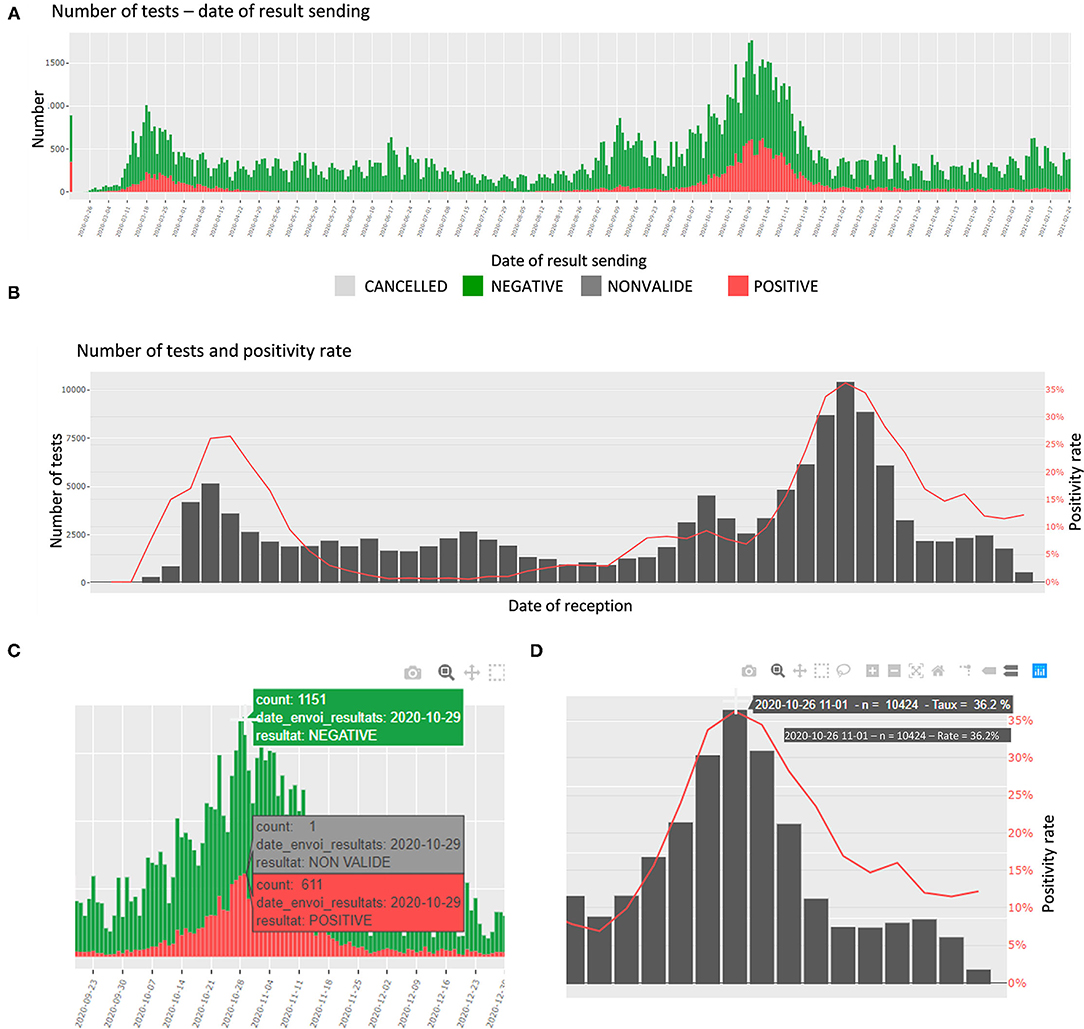
In this section, the number of tests and the proportions of daily positive specimens are detailed. The number of tests is presented per day of reception at the laboratory, per date of sending the results, and per age category. The proportions are presented per day and week of reception. In addition, an interactive table shows the number of tests per top categories of requesters. The plots are interactive to avoid cluttered visualization while allowing the user to choose the appropriate representation (zoom, labels, etc.) (Figures 2A–D).
|
Durations of the tests
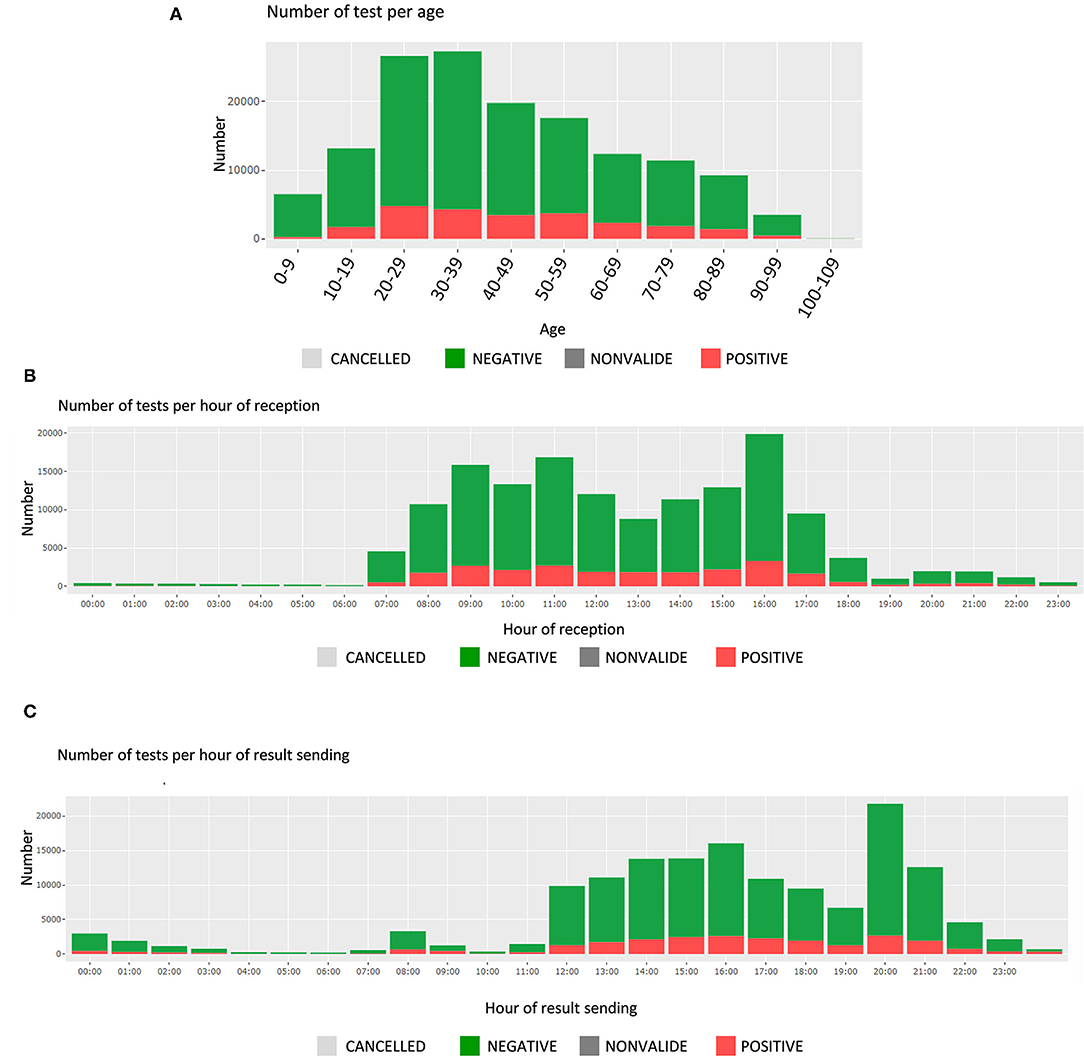
In this section, the user finds the time and date of test receipt and results sending, along with the associated test durations. These are presented by test results (positive or negative); the number of tests is displayed for each hour of a 24-hour day (Figures 3B,C). The average test durations and the average reception durations are displayed below for each reception and sending hourly period.
|
Viral load
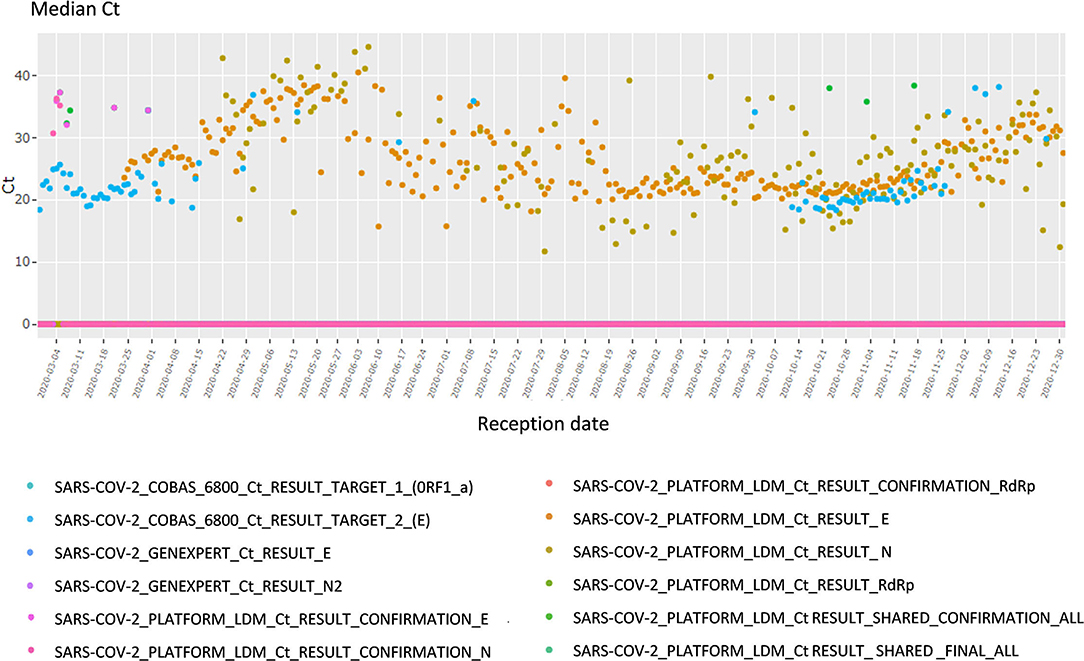
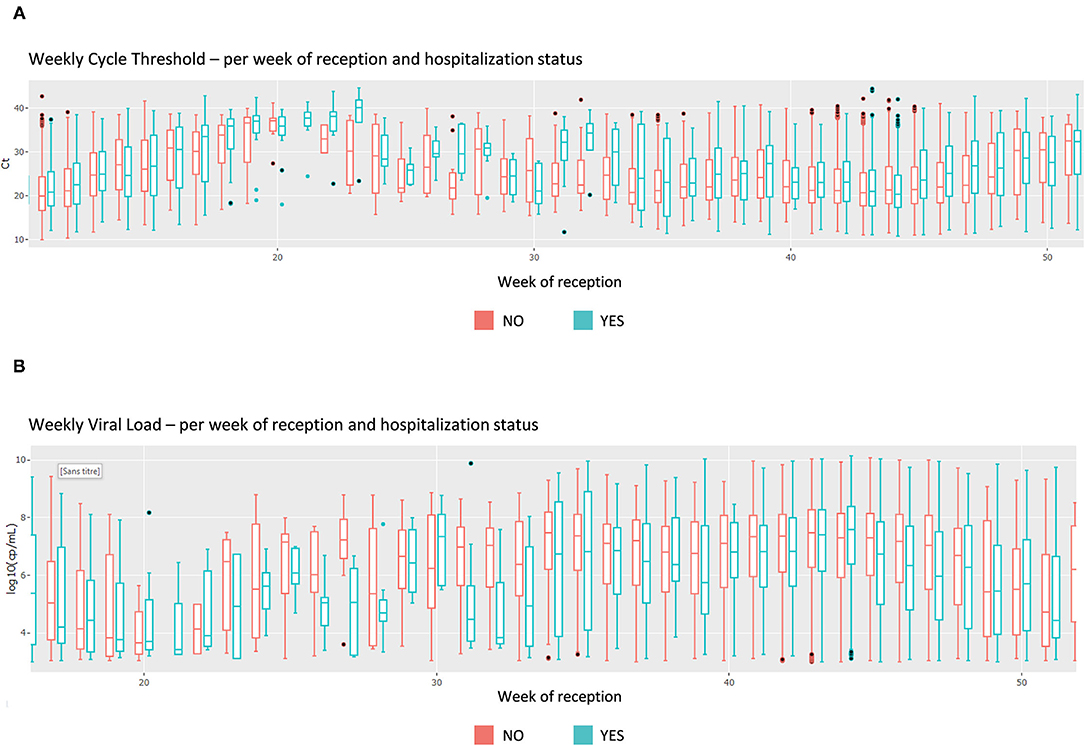
This section focuses on the viral load and cycle threshold (Ct), a KPI not presented in the first section. First, the median Ct for each day is shown, grouped by analysis (each device, each gene, each repetition); there is a total of 12 analyses (Figure 4). The viral load is also available in copy/millimeter (cp/mL) since April 9th, 2020, the date from which the laboratory started to keep this record. These are shown in a scatterplot crossed by type of sampling (blood, in nasopharyngeal secretions, etc.) as well as in a summary statistics table (Supplementary Figure 3).
|
NOT VALID
Finally, a descriptive table of the cases for which the result of the analysis was “NOT VALID” is displayed. The user can then identify any issue and investigate further. For sake of readability only some variables are shown (Supplementary Figure 4).
Comparison page
When moving to the Comparison page, the user sees first a global summary for all viruses. It shows the daily number of tests for each disease as well as the corresponding daily positivity rate. Then, like the Filter page, each virus information is shown in a specific tab. For each virus, the user can select the x-axis variable and the coloring variable. An error message is displayed when the chosen variables are the same (Table 1).
|
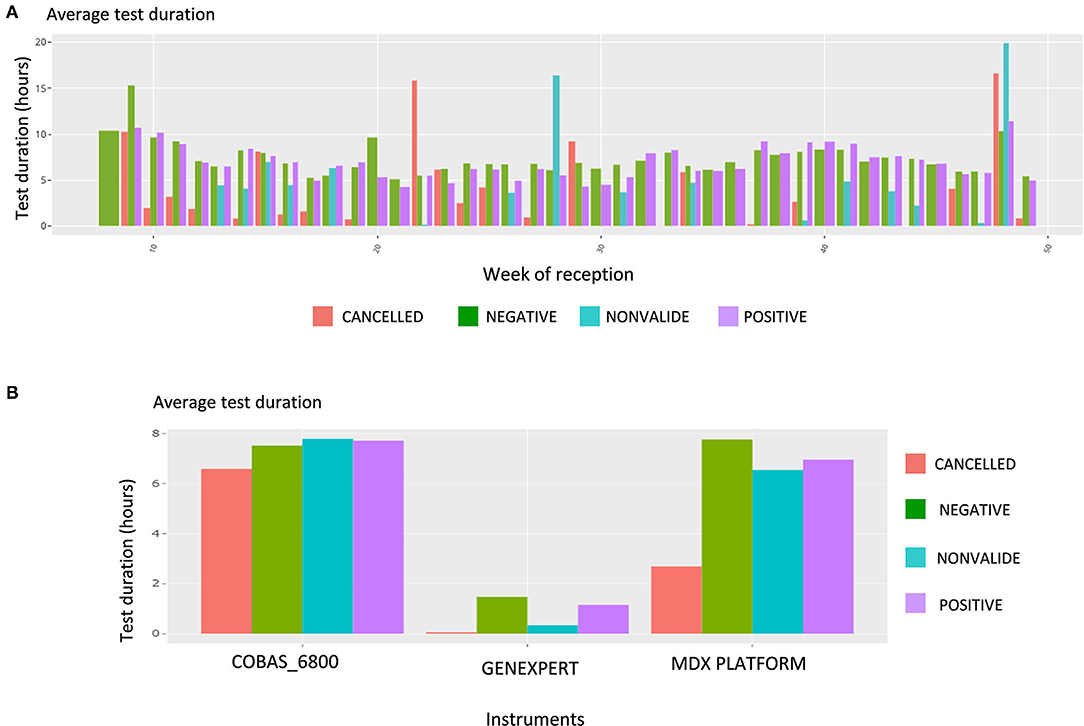
Upon this choice, the absolute and the relative number of tests are plotted, such as shown in Supplementary Figure 4 for gender and week of reception. The following plot shows data about the test duration. Figure 5 shows the average test duration per instrument, depending on the result of the test. This is especially useful to control the speed of some devices. Finally, the Ct information is displayed using boxplots (Figure 6A), a familiar representation to the end users. All the described figures are common to all viruses. For SARS-Cov-2, an additional boxplot of the viral load in log10 scale is displayed (Figure 6B). This additional boxplot appears only for this virus whose load can vary from 10,00 to 14,00,00,00,000 cp/mL. The log10 scale provides a more readable graph.
|
|
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. In some cases important information was missing from the references, and that information was added.