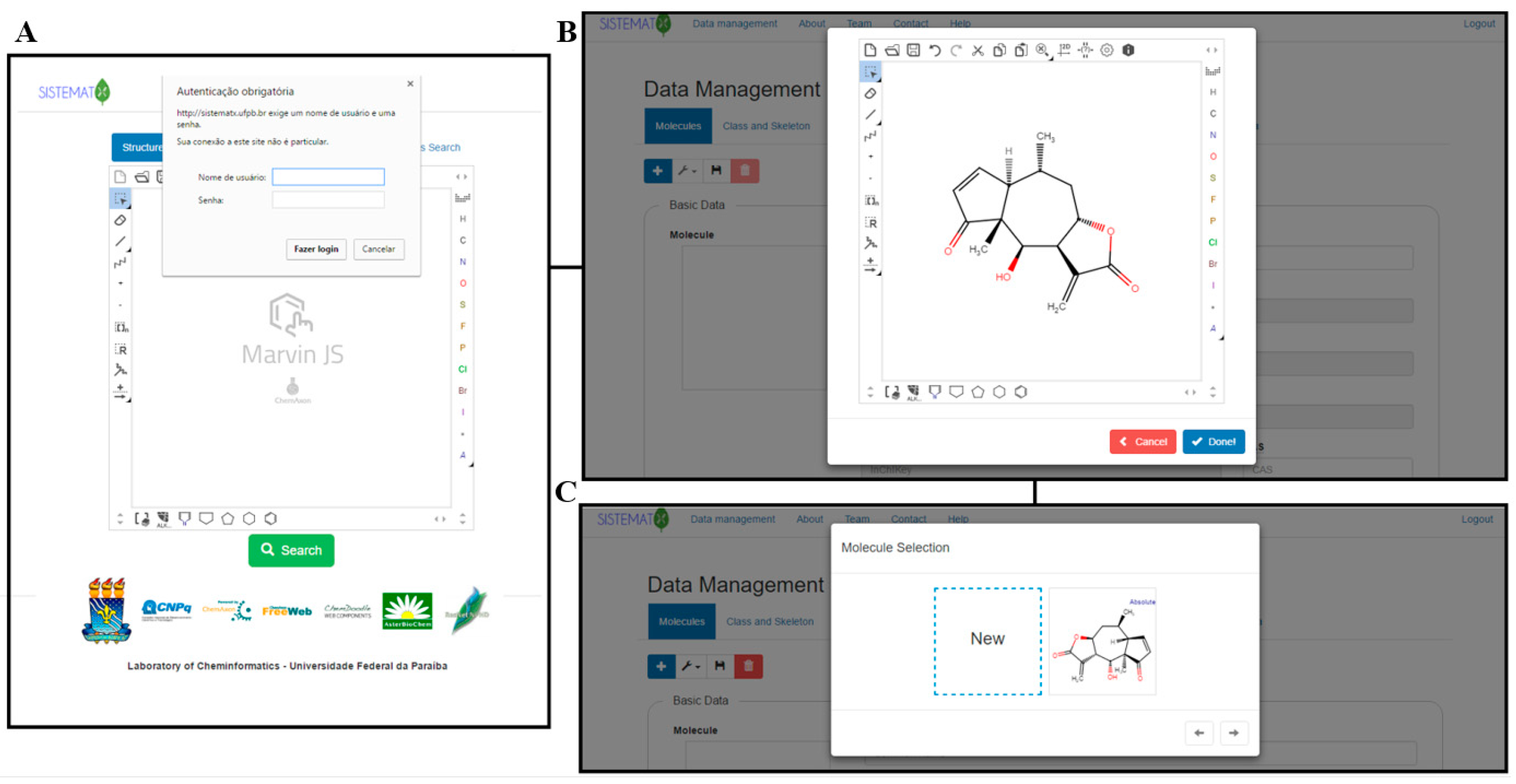
Difference between revisions of "Main Page/Featured article of the week/2018"
Shawndouglas (talk | contribs) (Added last week's article of the week.) |
Shawndouglas (talk | contribs) m (Text replacement - "\[\[LabArchives, LLC(.*)" to "[[Vendor:LabArchives, LLC$1") |
||
| (32 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{ombox | {{ombox | ||
| type = notice | | type = notice | ||
| text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - [[Main Page/Featured article of the week/2017|2017]] - 2018 | | text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - [[Main Page/Featured article of the week/2017|2017]] - 2018 - [[Main Page/Featured article of the week/2019|2019]] - [[Main Page/Featured article of the week/2020|2020]] - [[Main Page/Featured article of the week/2021|2021]] - [[Main Page/Featured article of the week/2022|2022]] - [[Main Page/Featured article of the week/2023|2023]] - [[Main Page/Featured article of the week/2024|2024]] | ||
}} | }} | ||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 18–24:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 17–31:</h2> | ||
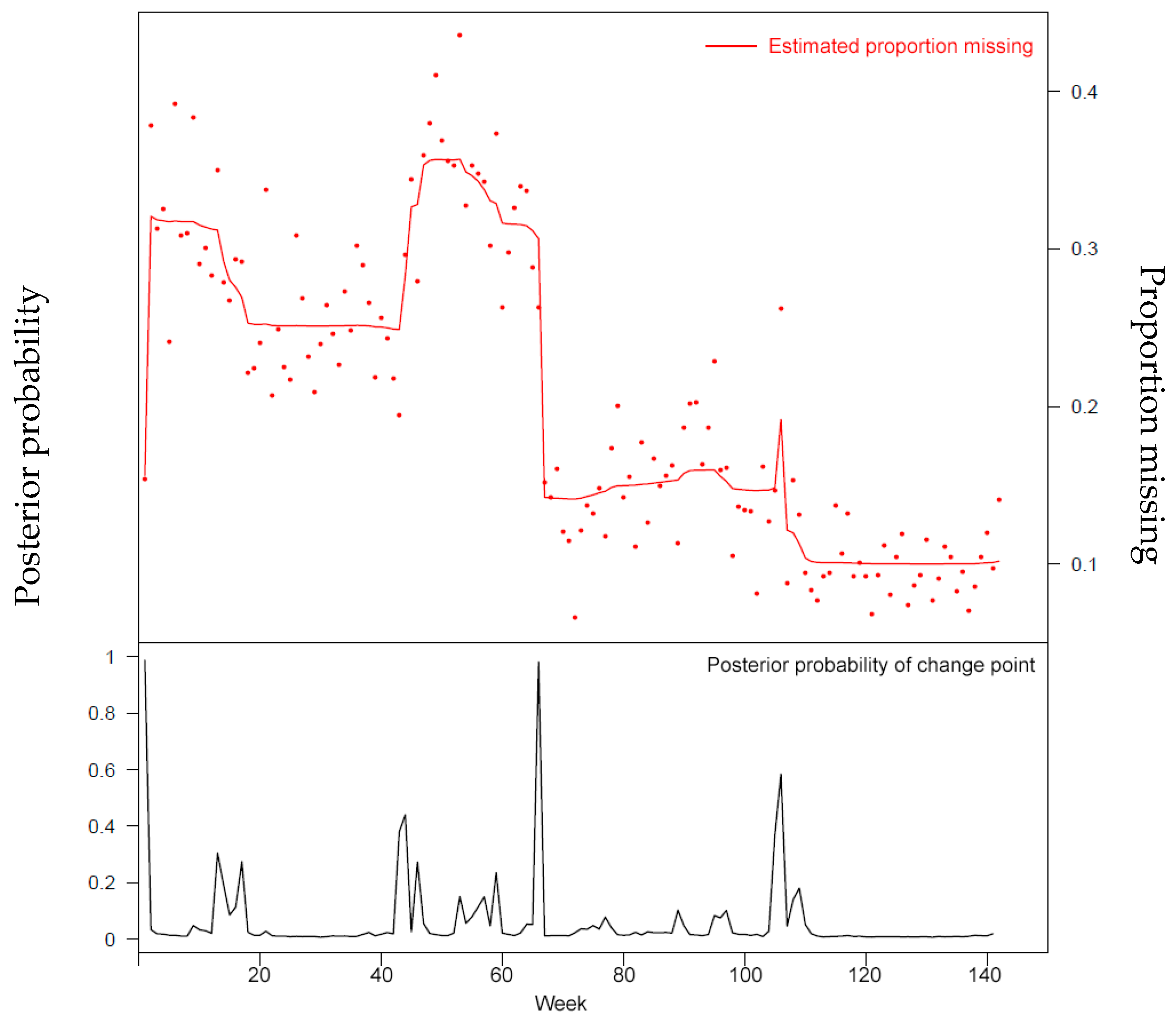
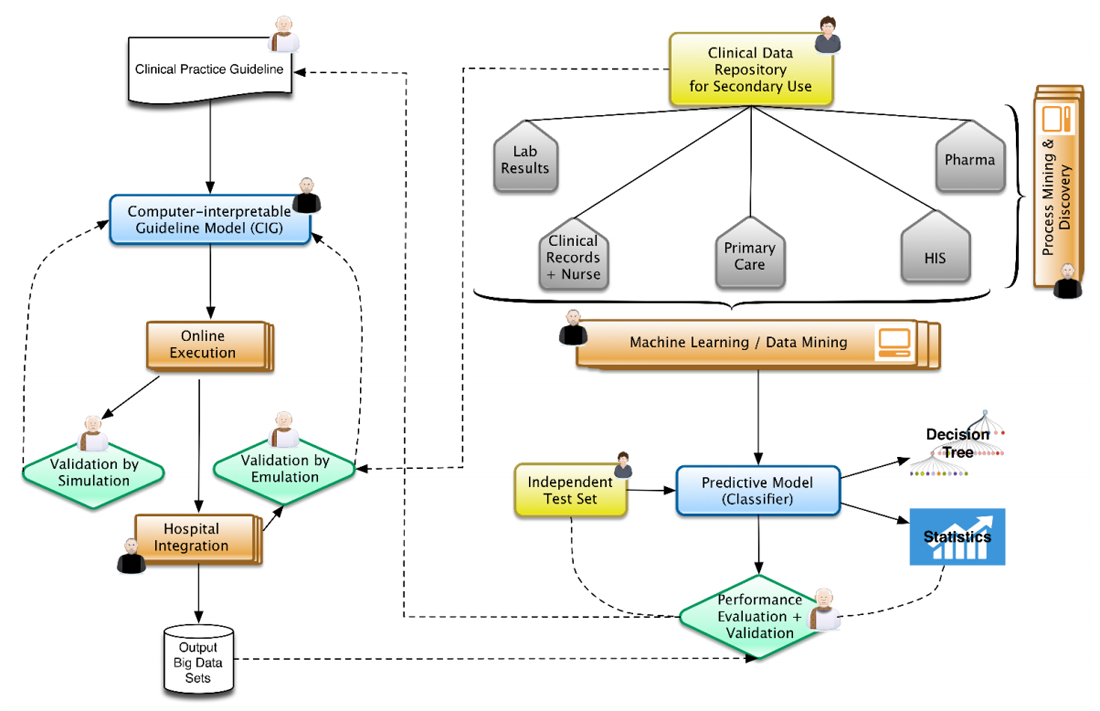
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Malykh JofHealthEng2018 2018.png|240px]]</div> | |||
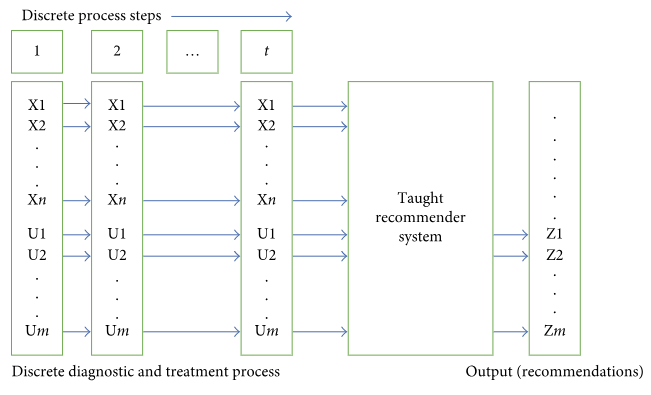
'''"[[Journal:Approaches to medical decision-making based on big clinical data|Approaches to medical decision-making based on big clinical data]]"''' | |||
The paper discusses different approaches to building a [[clinical decision support system]] based on big data. The authors sought to abstain from any data reduction and apply universal teaching and big data processing methods independent of disease classification standards. The paper assesses and compares the accuracy of recommendations among three options: case-based reasoning, simple single-layer neural network, and probabilistic neural network. Further, the paper substantiates the assumption regarding the most efficient approach to solving the specified problem. ('''[[Journal:Approaches to medical decision-making based on big clinical data|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 10–16:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Stura EpidemBiostatPubHealth2018 15-2.png|240px]]</div> | |||
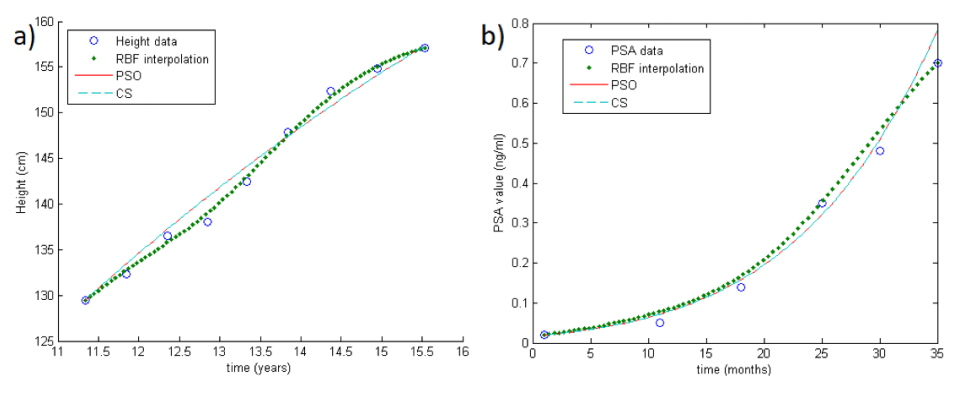
'''"[[Journal:A new numerical method for processing longitudinal data: Clinical applications|A new numerical method for processing longitudinal data: Clinical applications]]"''' | |||
Processing longitudinal data is a computational issue that arises in many applications, such as in aircraft design, medicine, optimal control, and weather forecasting. Given some longitudinal data, i.e., scattered measurements, the aim consists in approximating the parameters involved in the dynamics of the considered process. For this problem, a large variety of well-known methods have already been developed. Here, we propose an alternative approach to be used as an effective and accurate tool for the parameters fitting and prediction of individual trajectories from sparse longitudinal data. ('''[[Journal:A new numerical method for processing longitudinal data: Clinical applications|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 03–09:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Elaboudi AdvInBioinfo2018 2018.png|240px]]</div> | |||
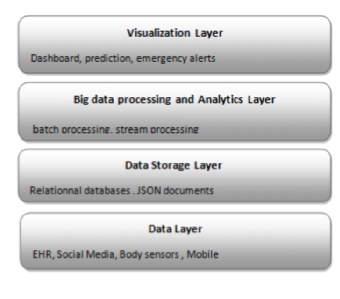
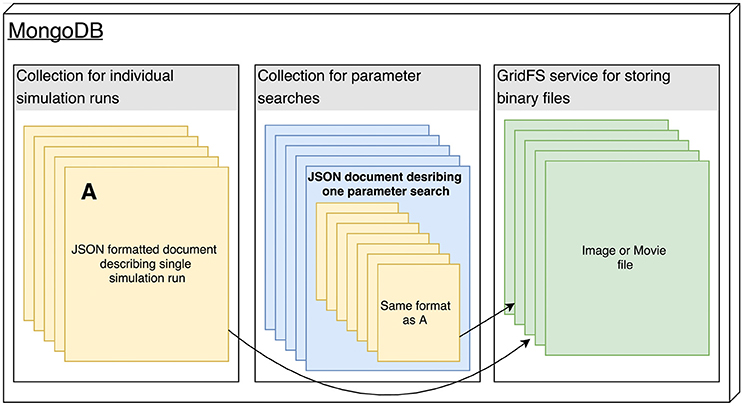
'''"[[Journal:Big data management for healthcare systems: Architecture, requirements, and implementation|Big data management for healthcare systems: Architecture, requirements, and implementation]]"''' | |||
The growing amount of data in the healthcare industry has made inevitable the adoption of big data techniques in order to improve the quality of healthcare delivery. Despite the integration of big data processing approaches and platforms in existing [[Information management|data management]] architectures for healthcare systems, these architectures face difficulties in preventing emergency cases. The main contribution of this paper is proposing an extensible big data architecture based on both stream computing and batch computing in order to enhance further the reliability of healthcare systems by generating real-time alerts and making accurate predictions on patient health condition. Based on the proposed architecture, a prototype implementation has been built for healthcare systems in order to generate real-time alerts. The suggested prototype is based on Spark and MongoDB tools. ('''[[Journal:Big data management for healthcare systems: Architecture, requirements, and implementation|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 26-December 02:</h2> | |||
'''"[[Journal:Support Your Data: A research data management guide for researchers|Support Your Data: A research data management guide for researchers]]"''' | |||
Researchers are faced with rapidly evolving expectations about how they should manage and share their data, code, and other [[research]] materials. To help them meet these expectations and generally manage and share their data more effectively, we are developing a suite of tools which we are currently referring to as "Support Your Data." These tools— which include a rubric designed to enable researchers to self-assess their current [[Information management|data management]] practices and a series of short guides which provide actionable [[information]] about how to advance practices as necessary or desired—are intended to be easily customizable to meet the needs of researchers working in a variety of institutional and disciplinary contexts. ('''[[Journal:Support Your Data: A research data management guide for researchers|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 19-25:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Fuertes GIMDS2018 7-1.png|240px]]</div> | |||
'''"[[Journal:CÆLIS: Software for assimilation, management, and processing data of an atmospheric measurement network|CÆLIS: Software for assimilation, management, and processing data of an atmospheric measurement network]]"''' | |||
Given the importance of atmospheric aerosols, the number of instruments and measurement networks which focus on its characterization is growing. Many challenges are derived from standardization of protocols, monitoring of instrument status to evaluate network [[Data integrity|data quality]], and manipulation and distribution of large volumes of data (raw and processed). CÆLIS is a software system which aims to simplify the management of a network, providing the scientific community a new tool for monitoring instruments, processing data in real time, and working with the data. Since 2008, CÆLIS has been successfully applied to the photometer calibration facility managed by the University of Valladolid, Spain, under the framework of the Aerosol Robotic Network (AERONET). Thanks to the use of advanced tools, this facility has been able to analyze a growing number of stations and data in real time, which greatly benefits network management and data quality control. The work describes the system architecture of CÆLIS and gives some examples of applications and data processing. ('''[[Journal:CÆLIS: Software for assimilation, management, and processing data of an atmospheric measurement network|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 12-18:</h2> | |||
'''"[[Journal:How could the ethical management of health data in the medical field inform police use of DNA?|How could the ethical management of health data in the medical field inform police use of DNA?]]"''' | |||
Various events paved the way for the production of ethical norms regulating biomedical practices, from the Nuremberg Code (1947)—produced by the international trial of Nazi regime leaders and collaborators—and the Declaration of Helsinki by the World Medical Association (1964) to the invention of the term “bioethics” by American biologist Van Rensselaer Potter. The ethics of biomedicine has given rise to various controversies—particularly in the fields of newborn screening, prenatal screening, and cloning—resulting in the institutionalization of ethical questions in the biomedical world of genetics. In 1994, France passed legislation (commonly known as the “bioethics laws”) to regulate medical practices in genetics. The medical community has also organized itself in order to manage ethical issues relating to its decisions, with a view to handling “practices with many strong uncertainties” and enabling clinical judgments and decisions to be made not by individual practitioners but rather by multidisciplinary groups drawing on different modes of judgment and forms of expertise. Thus, the biomedical approach to genetics has been characterized by various debates and the existence of public controversies. ('''[[Journal:Big data in the era of health information exchanges: Challenges and opportunities for public health|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 05-11:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Baseman Informatics2017 4-4.png|240px]]</div> | |||
'''"[[Journal:Big data in the era of health information exchanges: Challenges and opportunities for public health|Big data in the era of health information exchanges: Challenges and opportunities for public health]]"''' | |||
Public health surveillance of communicable diseases depends on timely, complete, accurate, and useful data that are collected across a number of health care and public health systems. [[Health information exchange]]s (HIEs) which support electronic sharing of data and [[information]] between health care organizations are recognized as a source of "big data" in health care and have the potential to provide public health with a single stream of data collated across disparate systems and sources. However, given these data are not collected specifically to meet public health objectives, it is unknown whether a public health agency’s (PHA’s) secondary use of the data is supportive of or presents additional barriers to meeting disease reporting and surveillance needs. To explore this issue, we conducted an assessment of big data that is available to a PHA—[[Public health laboratory|laboratory]] test results and clinician-generated notifiable condition report data—through its participation in an HIE. ('''[[Journal:Big data in the era of health information exchanges: Challenges and opportunities for public health|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 29-November 04:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Irawan ResIdeasOut2018 4.png|240px]]</div> | |||
'''"[[Journal:Promoting data sharing among Indonesian scientists: A proposal of a generic university-level research data management plan (RDMP)|Promoting data sharing among Indonesian scientists: A proposal of a generic university-level research data management plan (RDMP)]]"''' | |||
Every researcher needs data in their working ecosystem, but despite the resources (funding, time, and energy) they have spent to get the data, only a few are putting more real attention into [[Information management|data management]]. This paper mainly describes our recommendation of a research data management plan (RDMP) at the university level. This paper is an extension of our initiative, to be developed at the university or national level, while also in-line with current developments in scientific practices mandating data sharing and data re-use. Researchers can use this article as an assessment form to describe the setting of their research and data management. Researchers can also develop a more detailed RDMP to cater to a specific project's environment. ('''[[Journal:Promoting data sharing among Indonesian scientists: A proposal of a generic university-level research data management plan (RDMP)|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 22-28:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Backman BMCBio2016 17.gif|240px]]</div> | |||
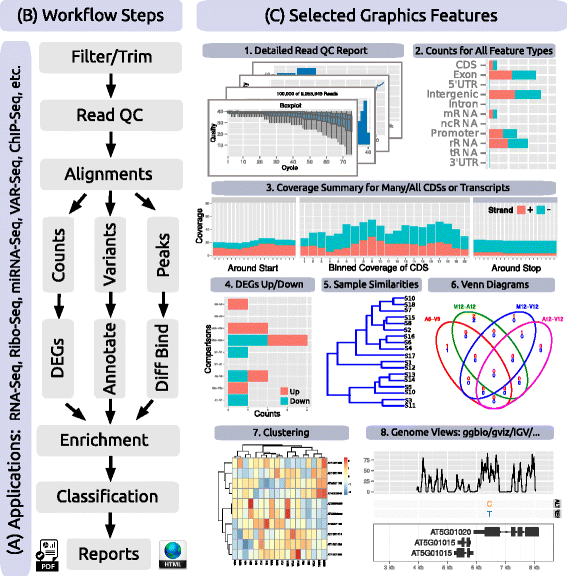
'''"[[Journal:systemPipeR: NGS workflow and report generation environment|systemPipeR: NGS workflow and report generation environment]]"''' | |||
Next-generation sequencing (NGS) has revolutionized how research is carried out in many areas of biology and medicine. However, the analysis of NGS data remains a major obstacle to the efficient utilization of the technology, as it requires complex multi-step processing of big data, demanding considerable computational expertise from users. While substantial effort has been invested on the development of software dedicated to the individual analysis steps of NGS experiments, insufficient resources are currently available for integrating the individual software components within the widely used R/Bioconductor environment into automated [[Workflow|workflows]] capable of running the analysis of most types of NGS applications from start-to-finish in a time-efficient and reproducible manner. ('''[[Journal:systemPipeR: NGS workflow and report generation environment|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 15-21:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Evans Informatics2017 4-4.png|240px]]</div> | |||
'''"[[Journal:A data quality strategy to enable FAIR, programmatic access across large, diverse data collections for high performance data analysis|A data quality strategy to enable FAIR, programmatic access across large, diverse data collections for high performance data analysis]]"''' | |||
To ensure seamless, programmatic access to data for high-performance computing (HPC) and [[Data analysis|analysis]] across multiple research domains, it is vital to have a methodology for standardization of both data and services. At the Australian National Computational Infrastructure (NCI) we have developed a data quality strategy (DQS) that currently provides processes for: (1) consistency of data structures needed for a high-performance data (HPD) platform; (2) [[quality control]] (QC) through compliance with recognized community standards; (3) benchmarking cases of operational performance tests; and (4) [[quality assurance]] (QA) of data through demonstrated functionality and performance across common platforms, tools, and services. ('''[[Journal:A data quality strategy to enable FAIR, programmatic access across large, diverse data collections for high performance data analysis|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 08-14:</h2> | |||
'''"[[Journal:How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology|How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology]]"''' | |||
Big data and comparative effectiveness research methodologies can be applied within the framework of a rapid-learning health care system (RLHCS) to accelerate discovery and to help turn the dream of fully personalized medicine into a reality. We synthesize recent advances in [[genomics]] with trends in big data to provide a forward-looking perspective on the potential of new advances to usher in an era of personalized radiation therapy, with emphases on the power of RLHCS to accelerate discovery and the future of individualized radiation treatment planning. ('''[[Journal:How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 01-07:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig5 eSilva Sensors2018 18-8.jpg|240px]]</div> | |||
'''"[[Journal:Wireless positioning in IoT: A look at current and future trends|Wireless positioning in IoT: A look at current and future trends]]"''' | |||
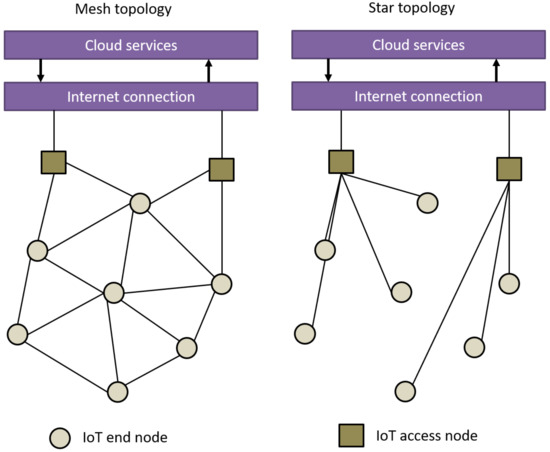
Connectivity solutions for the [[internet of things]] (IoT) aim to support the needs imposed by several applications or use cases across multiple sectors, such as logistics, [[Agriculture industry|agriculture]], asset management, or smart lighting. Each of these applications has its own challenges to solve, such as dealing with large or massive networks, low and ultra-low latency requirements, long battery life requirements (i.e., more than ten years operation on battery), continuously monitoring of the location of certain nodes, security, and authentication. Hence, a part of picking a connectivity solution for a certain application depends on how well its features solve the specific needs of the end application. One key feature that we see as a need for future IoT networks is the ability to provide location-based [[information]] for large-scale IoT applications. ('''[[Journal:Wireless positioning in IoT: A look at current and future trends|Full article...]]''')<br /> | |||
|- | |||
|<br /> | |||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 24-30:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Mahlaola SAJouBioLaw2017 10-2.png|240px]]</div> | |||
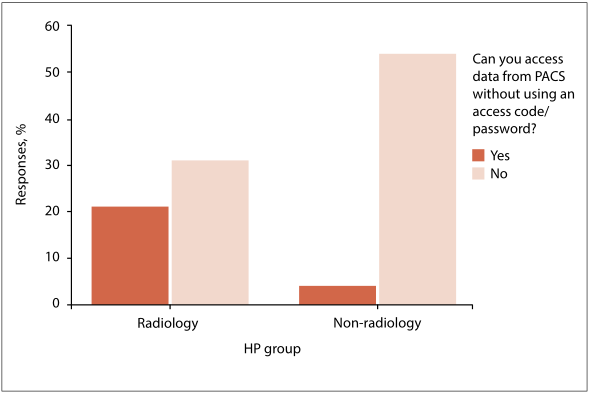
'''"[[Journal:Password compliance for PACS work stations: Implications for emergency-driven medical environments|Password compliance for PACS work stations: Implications for emergency-driven medical environments]]"''' | |||
The effectiveness of password usage in data security remains an area of high scrutiny. Literature findings do not inspire confidence in the use of passwords. Human factors such as the acceptance of and compliance with minimum standards of data security are considered significant determinants of effective data-security practices. However, human and technical factors alone do not provide solutions if they exclude the context in which the technology is applied. | |||
Objectives: To reflect on the outcome of a dissertation which argues that the minimum standards of effective password use prescribed by the [[information]] security sector are not suitable to the emergency-driven medical environment, and that their application as required by law raises new and unforeseen ethical dilemmas. ('''[[Journal:Password compliance for PACS work stations: Implications for emergency-driven medical environments|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 10-23:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig5 Kayser TechInnoManRev2018 8-3.png|240px]]</div> | |||
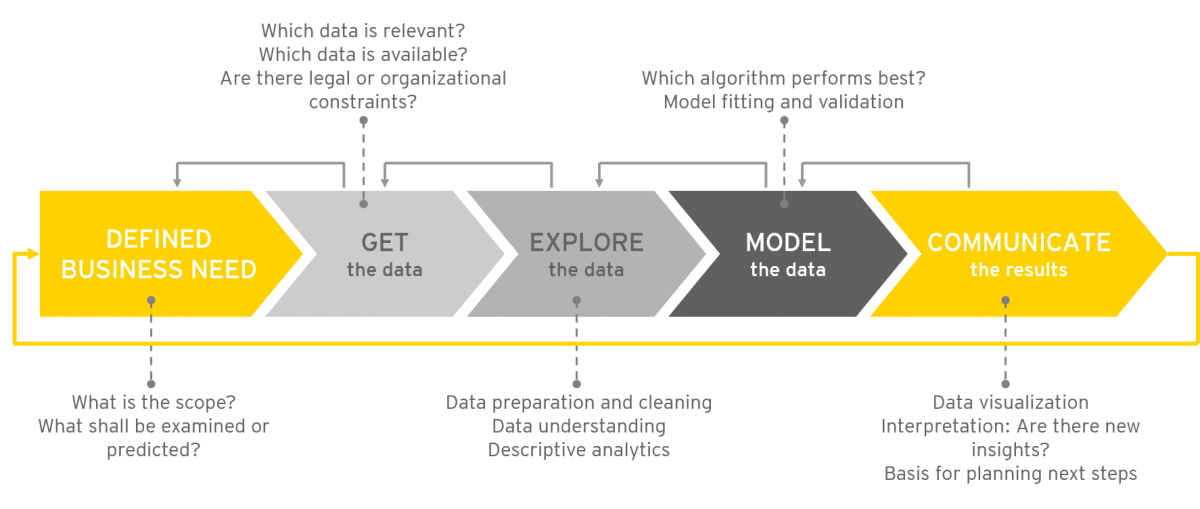
'''"[[Journal:Data science as an innovation challenge: From big data to value proposition|Data science as an innovation challenge: From big data to value proposition]]"''' | |||
Analyzing “big data” holds huge potential for generating business value. The ongoing advancement of tools and technology over recent years has created a new ecosystem full of opportunities for data-driven innovation. However, as the amount of available data rises to new heights, so too does complexity. Organizations are challenged to create the right contexts, by shaping interfaces and processes, and by asking the right questions to guide the [[data analysis]]. Lifting the innovation potential requires teaming and focus to efficiently assign available resources to the most promising initiatives. With reference to the innovation process, this article will concentrate on establishing a process for analytics projects from first ideas to realization (in most cases, a running application). The question we tackle is: what can the practical discourse on big data and analytics learn from innovation management? ('''[[Journal:Data science as an innovation challenge: From big data to value proposition|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 3-9:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Murtagh BigDataCogComp2018 2-2.jpg|240px]]</div> | |||
'''"[[Journal:The development of data science: Implications for education, employment, research, and the data revolution for sustainable development|The development of data science: Implications for education, employment, research, and the data revolution for sustainable development]]"''' | |||
In data science, we are concerned with the integration of relevant sciences in observed and empirical contexts. This results in the unification of analytical methodologies, and of observed and empirical data contexts. Given the dynamic nature of convergence, the origins and many evolutions of the data science theme are described. The following are covered in this article: the rapidly growing post-graduate university course provisioning for data science; a preliminary study of employability requirements; and how past eminent work in the social sciences and other areas, certainly mathematics, can be of immediate and direct relevance and benefit for innovative methodology, and for facing and addressing the ethical aspect of big data [[Data analysis|analytics]], relating to data aggregation and scale effects. ('''[[Journal:The development of data science: Implications for education, employment, research, and the data revolution for sustainable development|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 27–September 2:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Leroux Agri2018 8-6.jpg|240px]]</div> | |||
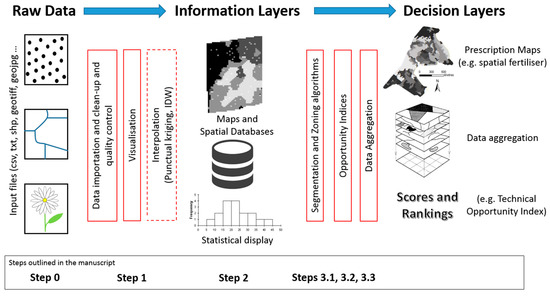
'''"[[Journal:GeoFIS: An open-source decision support tool for precision agriculture data|GeoFIS: An open-source decision support tool for precision agriculture data]]"''' | |||
The world we live in is an increasingly spatial and temporal data-rich environment, and the [[agriculture industry]] is no exception. However, data needs to be processed in order to first get [[information]] and then make informed management decisions. The concepts of "precision agriculture" and "smart agriculture" can and will be fully effective when methods and tools are available to practitioners to support this transformation. An open-source program called GeoFIS has been designed with this objective. It was designed to cover the whole process from spatial data to spatial information and decision support. The purpose of this paper is to evaluate the abilities of GeoFIS along with its embedded algorithms to address the main features required by farmers, advisors, or spatial analysts when dealing with precision agriculture data. Three case studies are investigated in the paper: (i) mapping of the spatial variability in the data, (ii) evaluation and cross-comparison of the opportunity for site-specific management in multiple fields, and (iii) delineation of within-field zones for variable-rate applications when these latter are considered opportune. ('''[[Journal:GeoFIS: An open-source decision support tool for precision agriculture data|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 20–26:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 BezuidenhoutDataSciJo2017 16.png|240px]]</div> | |||
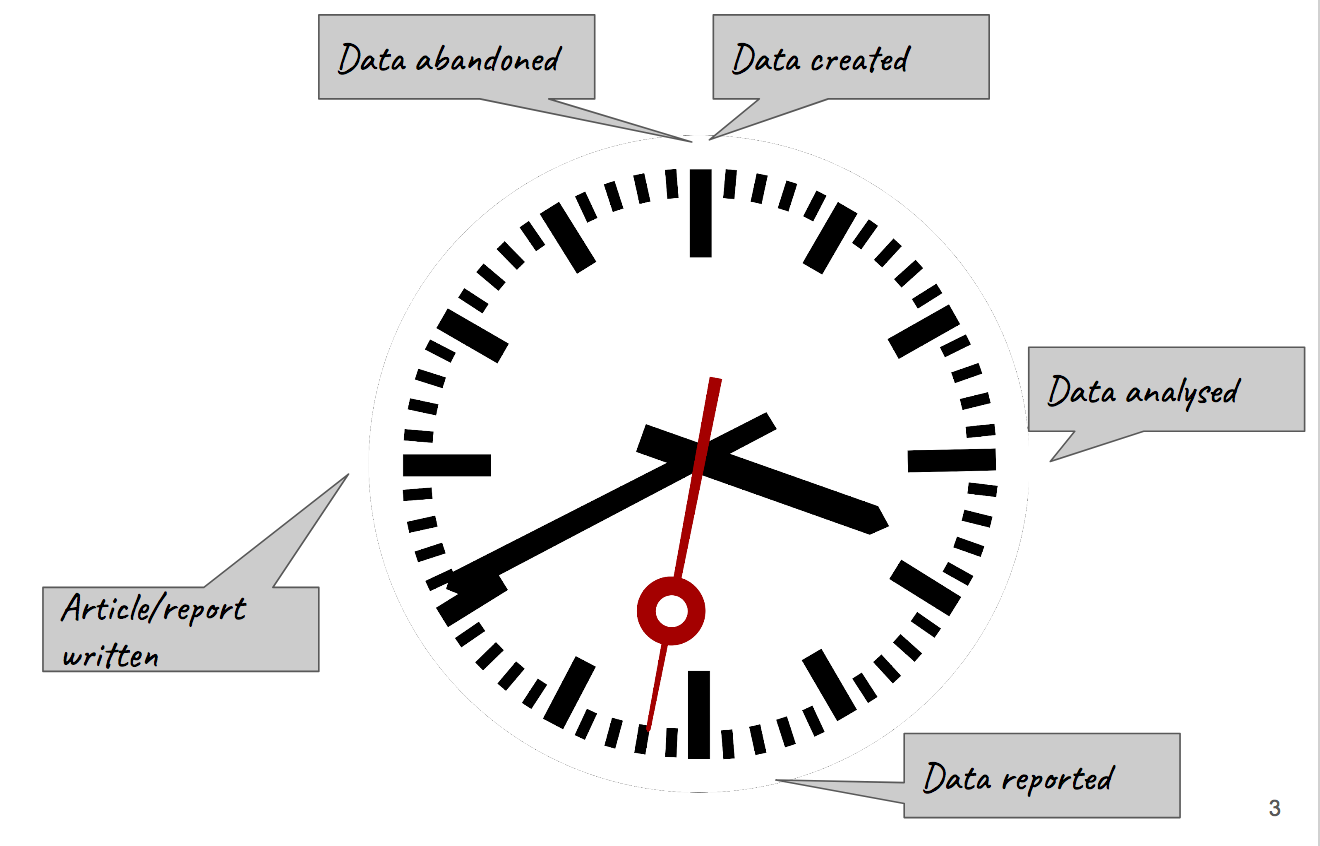
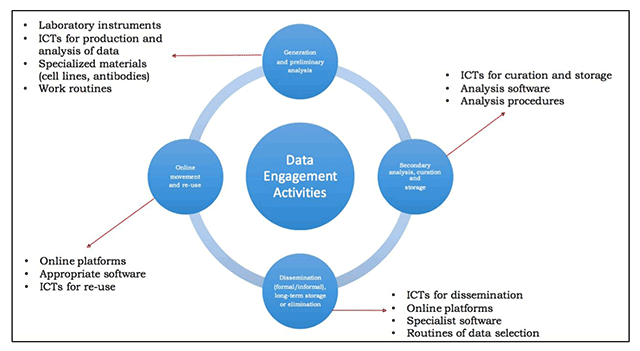
'''"[[Journal:Technology transfer and true transformation: Implications for open data|Technology transfer and true transformation: Implications for open data]]"''' | |||
When considering the “openness” of data, it is unsurprising that most conversations focus on the online environment—how data is collated, moved, and recombined for multiple purposes. Nonetheless, it is important to recognize that the movements online are only part of the data lifecycle. Indeed, considering where and how data are created—namely, the research setting—are of key importance to open data initiatives. In particular, such insights offer key understandings of how and why scientists engage with in practices of openness, and how data transitions from personal control to public ownership. This paper examines research settings in low/middle-income countries (LMIC) to better understand how resource limitations influence open data buy-in. ('''[[Journal:Technology transfer and true transformation: Implications for open data|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 13–19:</h2> | |||
'''"[[Journal:Eleven quick tips for architecting biomedical informatics workflows with cloud computing|Eleven quick tips for architecting biomedical informatics workflows with cloud computing]]"''' | |||
[[Cloud computing]] has revolutionized the development and operations of hardware and software across diverse technological arenas, yet academic biomedical research has lagged behind despite the numerous and weighty advantages that cloud computing offers. Biomedical researchers who embrace cloud computing can reap rewards in cost reduction, decreased development and maintenance workload, increased reproducibility, ease of sharing data and software, enhanced security, horizontal and vertical scalability, high availability, a thriving technology partner ecosystem, and much more. Despite these advantages that cloud-based [[workflow]]s offer, the majority of scientific software developed in academia does not utilize cloud computing and must be migrated to the cloud by the user. In this article, we present 11 quick tips for designing biomedical informatics workflows on compute clouds, distilling knowledge gained from experience developing, operating, maintaining, and distributing software and virtualized appliances on the world’s largest cloud. Researchers who follow these tips stand to benefit immediately by migrating their workflows to cloud computing and embracing the paradigm of abstraction. ('''[[Journal:Eleven quick tips for architecting biomedical informatics workflows with cloud computing|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 6–12:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig4 SprengholzQuantMethSci2018 14-2.png|240px]]</div> | |||
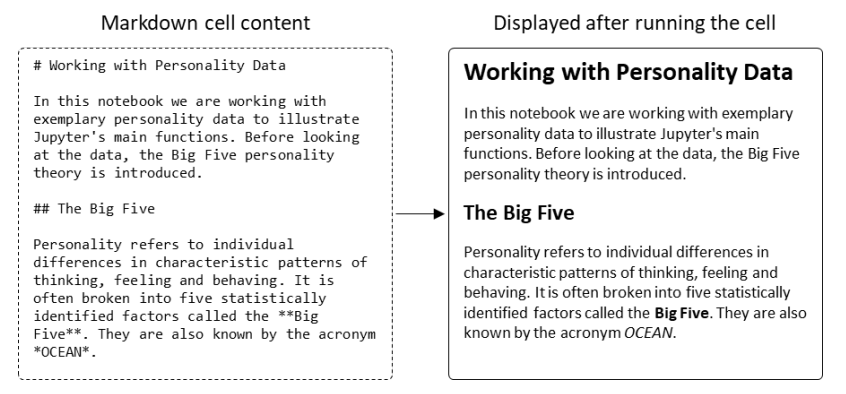
'''"[[Journal:Welcome to Jupyter: Improving collaboration and reproduction in psychological research by using a notebook system|Welcome to Jupyter: Improving collaboration and reproduction in psychological research by using a notebook system]]"''' | |||
The reproduction of findings from psychological research has been proven difficult. Abstract description of the data analysis steps performed by researchers is one of the main reasons why reproducing or even understanding published findings is so difficult. With the introduction of [[Jupyter Notebook]], a new tool for the organization of both static and dynamic [[information]] became available. The software allows blending explanatory content like written text or images with code for preprocessing and analyzing scientific data. Thus, Jupyter helps document the whole research process from ideation over data analysis to the interpretation of results. This fosters both collaboration and scientific quality by helping researchers to organize their work. This tutorial is an introduction to Jupyter. It explains how to set up and use the notebook system. While introducing its key features, the advantages of using Jupyter Notebook for psychological research become obvious. ('''[[Journal:Welcome to Jupyter: Improving collaboration and reproduction in psychological research by using a notebook system|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 30–August 5:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig8 ErgüzenAppSci2018 8-6.png|240px]]</div> | |||
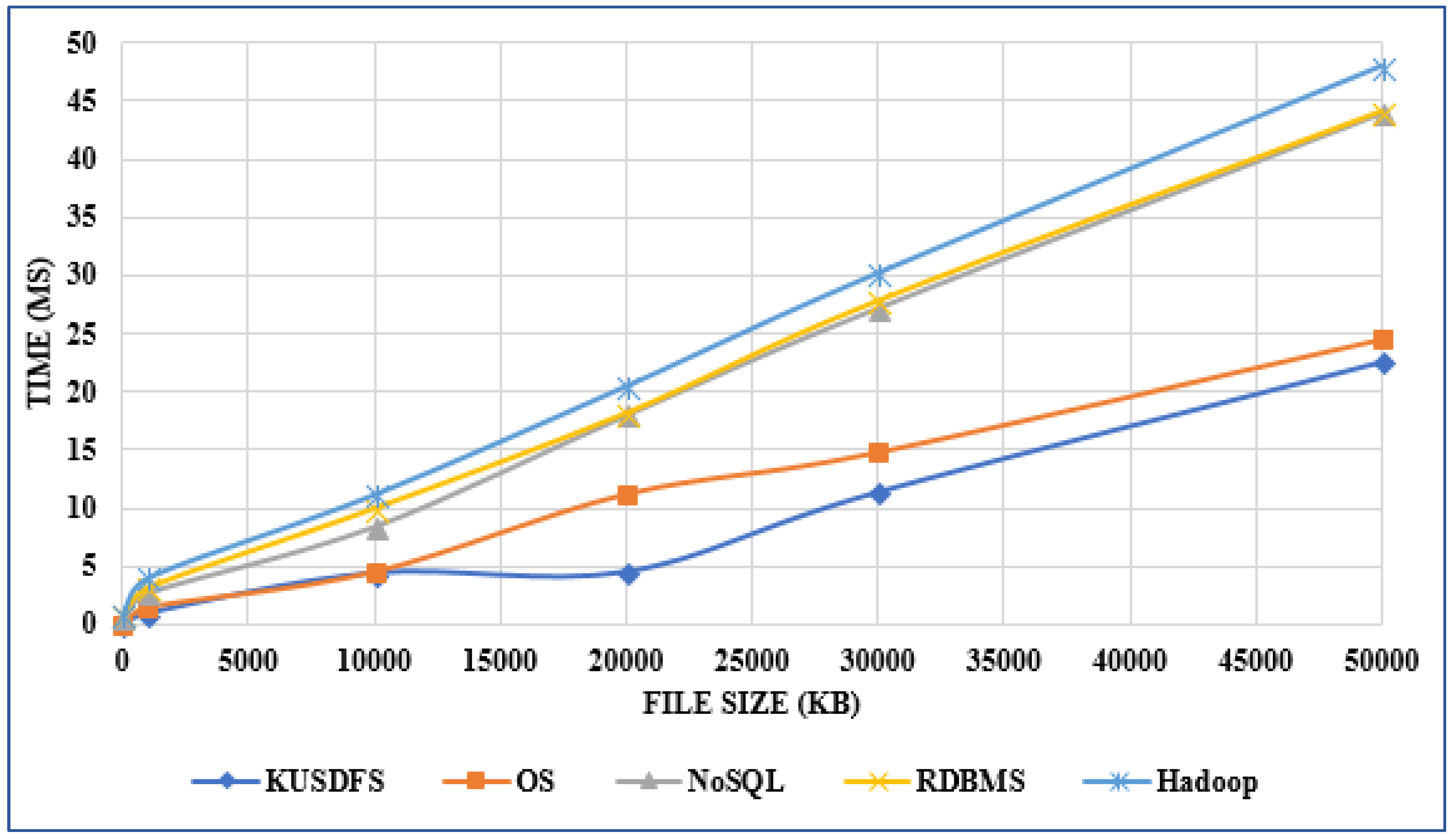
'''"[[Journal:Developing a file system structure to solve healthcare big data storage and archiving problems using a distributed file system|Developing a file system structure to solve healthcare big data storage and archiving problems using a distributed file system]]"''' | |||
Recently, the use of the internet has become widespread, increasing the use of mobile phones, tablets, computers, internet of things (IoT) devices, and other digital sources. In the healthcare sector, with the help of next generation digital medical equipment, this digital world also has tended to grow in an unpredictable way such that nearly 10 percent of global data is healthcare-related, continuing to grow beyond what other sectors have. This progress has greatly enlarged the amount of produced data which cannot be resolved with conventional methods. In this work, an efficient model for the storage of medical images using a distributed file system structure has been developed. With this work, a robust, available, scalable, and serverless solution structure has been produced, especially for storing large amounts of data in the medical field. Furthermore, the security level of the system is extreme by use of static Internet Protocol (IP) addresses, user credentials, and synchronously encrypted file contents. ('''[[Journal:Developing a file system structure to solve healthcare big data storage and archiving problems using a distributed file system|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 23–29:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 BaldominosIntJOfIMAI2018 4-7.png|240px]]</div> | |||
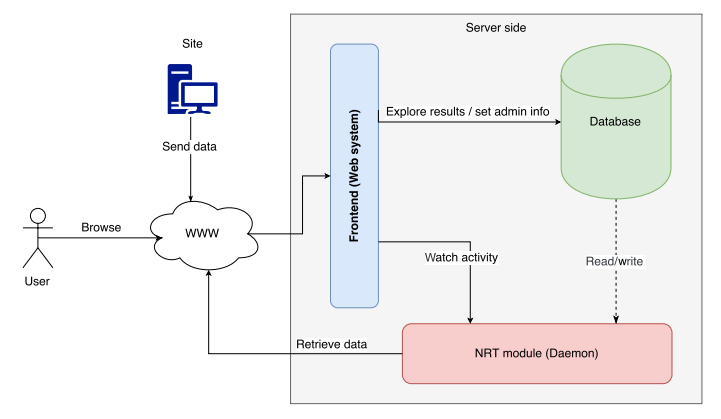
'''"[[Journal:DataCare: Big data analytics solution for intelligent healthcare management|DataCare: Big data analytics solution for intelligent healthcare management]]"''' | |||
This paper presents DataCare, a solution for intelligent healthcare management. This product is able not only to retrieve and aggregate data from different key performance indicators in healthcare centers, but also to estimate future values for these key performance indicators and, as a result, fire early alerts when undesirable values are about to occur or provide recommendations to improve the quality of service. DataCare’s core processes are built over a free and open-source cross-platform document-oriented database (MongoDB), and Apache Spark, an open-source cluster computing framework. This architecture ensures high scalability capable of processing very high data volumes coming at rapid speeds from a large set of sources. This article describes the architecture designed for this project and the results obtained after conducting a pilot in a healthcare center. Useful conclusions have been drawn regarding how key performance indicators change based on different situations, and how they affect patients’ satisfaction. ('''[[Journal:DataCare: Big data analytics solution for intelligent healthcare management|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 16–22:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Kalathil FrontInResMetAnal2018 2.jpg|240px]]</div> | |||
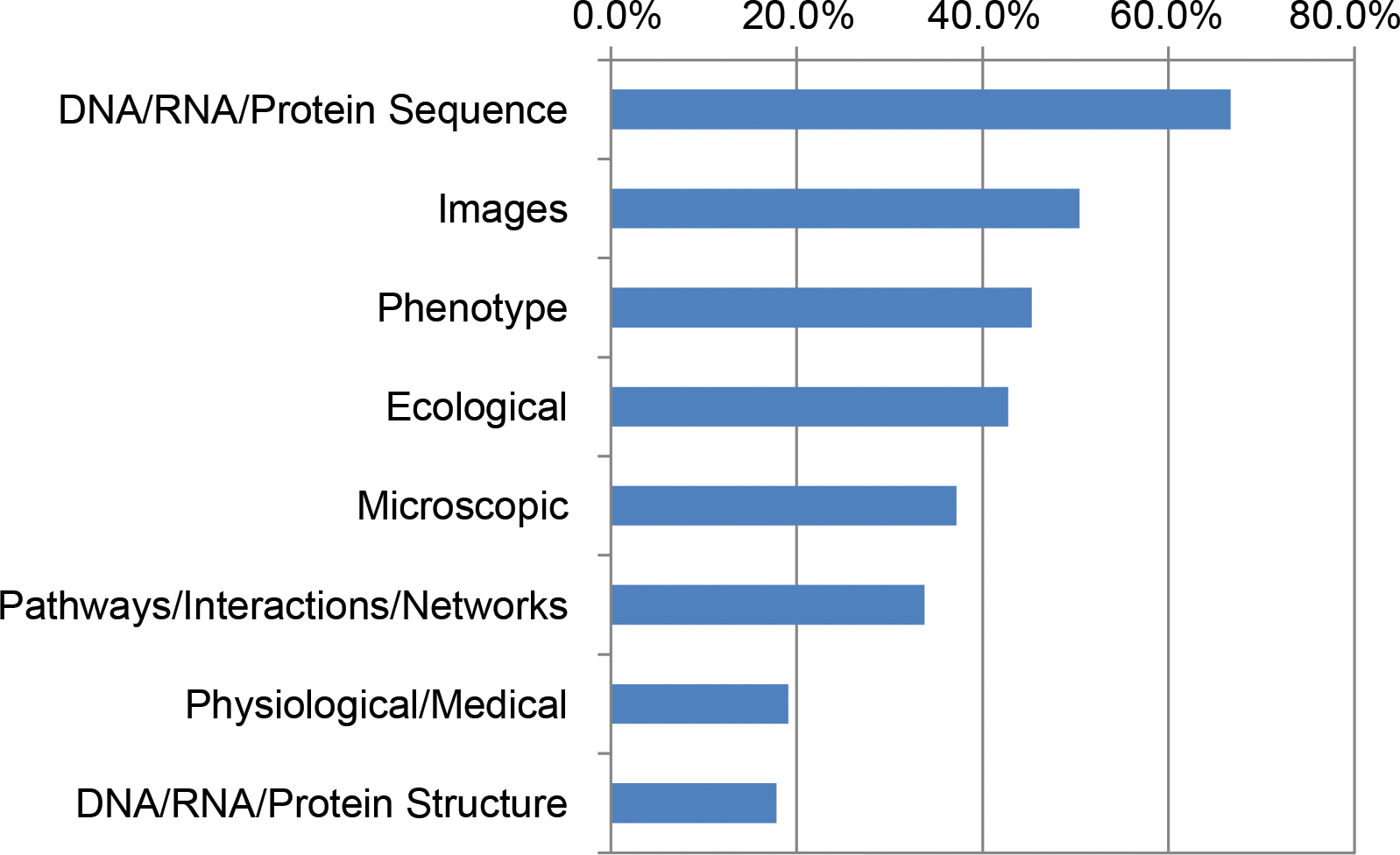
'''"[[Journal:Application of text analytics to extract and analyze material–application pairs from a large scientific corpus|Application of text analytics to extract and analyze material–application pairs from a large scientific corpus]]"''' | |||
When assessing the importance of materials (or other components) to a given set of applications, machine analysis of a very large corpus of scientific abstracts can provide an analyst a base of insights to develop further. The use of text analytics reduces the time required to conduct an evaluation, while allowing analysts to experiment with a multitude of different hypotheses. Because the scope and quantity of [[metadata]] analyzed can, and should, be large, any divergence from what a human analyst determines and what the text analysis shows provides a prompt for the human analyst to reassess any preliminary findings. In this work, we have successfully extracted material–application pairs and ranked them on their importance. This method provides a novel way to map scientific advances in a particular material to the application for which it is used. ('''[[Journal:Application of text analytics to extract and analyze material–application pairs from a large scientific corpus|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 9–15:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig6 BuřitaJOfSysInteg2018 9-1.png|240px]]</div> | |||
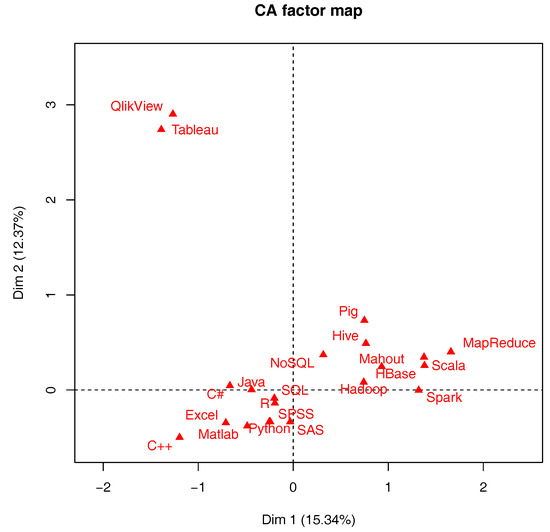
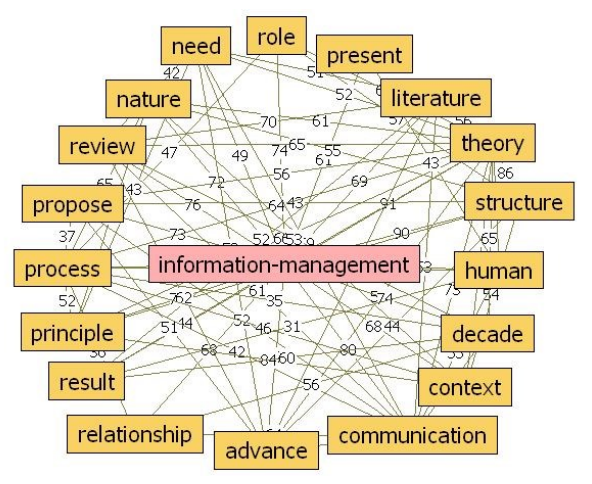
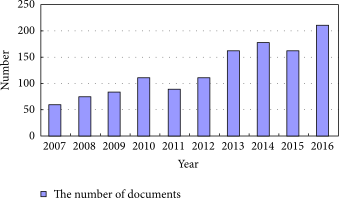
'''"[[Journal:Information management in context of scientific disciplines|Information management in context of scientific disciplines]]"''' | |||
This paper aims to analyze publications with the theme of [[information management]] (IM), cited on Web of Science (WoS) or Scopus. The frequency of publishing about IM has approached linear growth, from a few articles in the period 1966–1970 to 100 at the WoS and 600 at Scopus in the period 2011–2015. From this selection of publications, this analysis looked at 21 of the most cited articles on WoS and 21 of the most cited articles on Scopus, published in 31 different journals, oriented to [[informatics]] and computer science; economics, business, and management; medicine and psychology; art and the humanities; and ergonomics. The diversity of interest in IM in various areas of science, technology, and practice was confirmed. The content of the selected articles was analyzed in its area of interest, in relation to IM, and whether the definition of IM was mentioned. One of the goals was to confirm the hypothesis that IM is included in many scientific disciplines, that the concept of IM is used loosely, and it is mostly mentioned as part of data or information processing. ('''[[Journal:Information management in context of scientific disciplines|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 2–8:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 CaoProcess2018 6-5.png|240px]]</div> | |||
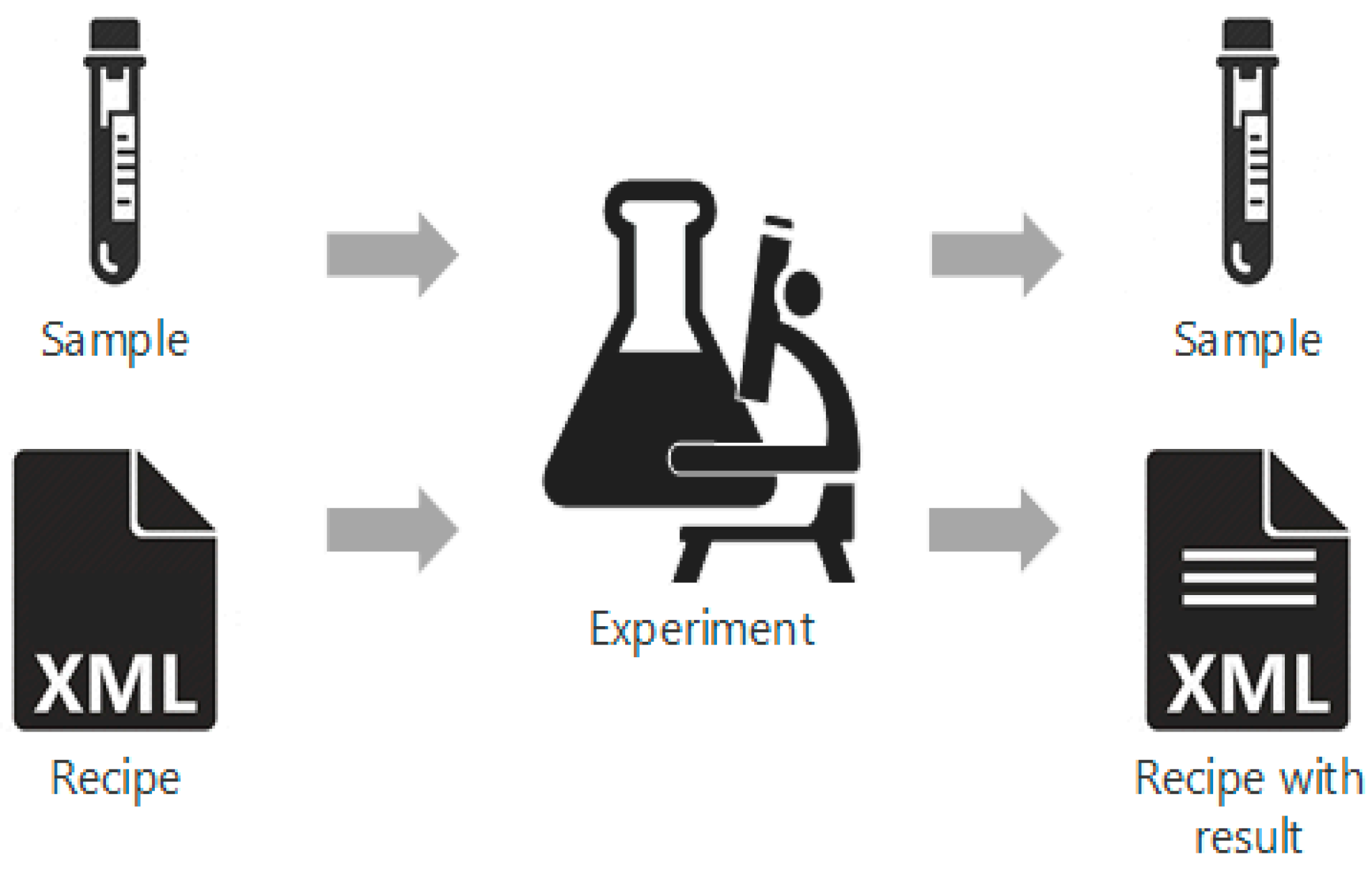
'''"[[Journal:A systematic framework for data management and integration in a continuous pharmaceutical manufacturing processing line|A systematic framework for data management and integration in a continuous pharmaceutical manufacturing processing line]]"''' | |||
As the pharmaceutical industry seeks more efficient methods for the production of higher value therapeutics, the associated [[data analysis]], [[data visualization]], and predictive modeling require dependable data origination, management, transfer, and integration. As a result, the management and integration of data in a consistent, organized, and reliable manner is a big challenge for the pharmaceutical industry. In this work, an ontological [[information]] infrastructure is developed to integrate data within manufacturing plants and analytical [[Laboratory|laboratories]]. The ANSI/ISA-88 batch control standard has been adapted in this study to deliver a well-defined data structure that will improve the data communication inside the system architecture for continuous processing. All the detailed information of the lab-based experiment and process manufacturing—including equipment, samples, and parameters—are documented in the recipe. This recipe model is implemented into a process control system (PCS), data historian, and [[electronic laboratory notebook]] (ELN). ('''[[Journal:A systematic framework for data management and integration in a continuous pharmaceutical manufacturing processing line|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 25–July 1:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 BaronePLOSCompBio2017 13-11.png|240px]]</div> | |||
'''"[[Journal:Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators|Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators]]"''' | |||
In a 2016 survey of 704 National Science Foundation (NSF) Biological Sciences Directorate principal investigators (BIO PIs), nearly 90% indicated they are currently or will soon be analyzing large data sets. BIO PIs considered a range of computational needs important to their work, including high-performance computing (HPC), [[bioinformatics]] support, multistep workflows, updated analysis software, and the ability to store, share, and publish data. Previous studies in the United States and Canada emphasized infrastructure needs. However, BIO PIs said the most pressing unmet needs are training in data integration, data management, and scaling analyses for HPC, acknowledging that data science skills will be required to build a deeper understanding of life. This portends a growing data knowledge gap in biology and challenges institutions and funding agencies to redouble their support for computational training in biology. ('''[[Journal:Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators|Full article...]]''')<br /> | |||
|- | |||
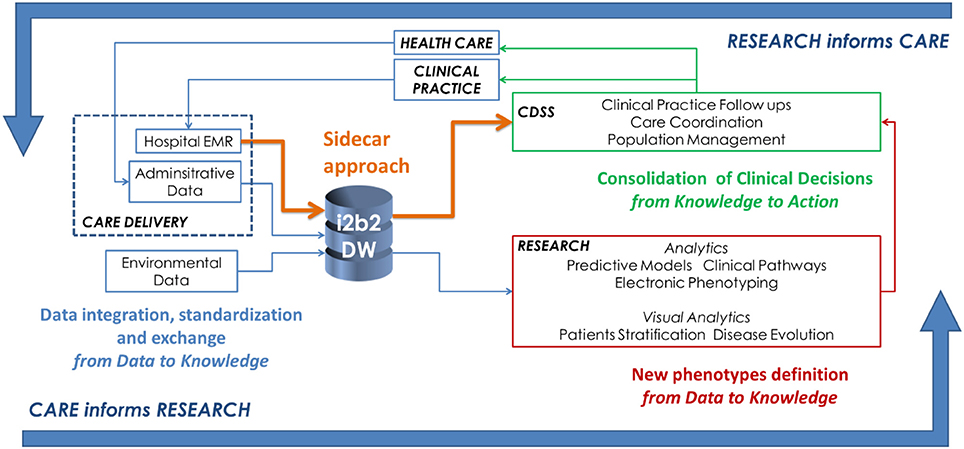
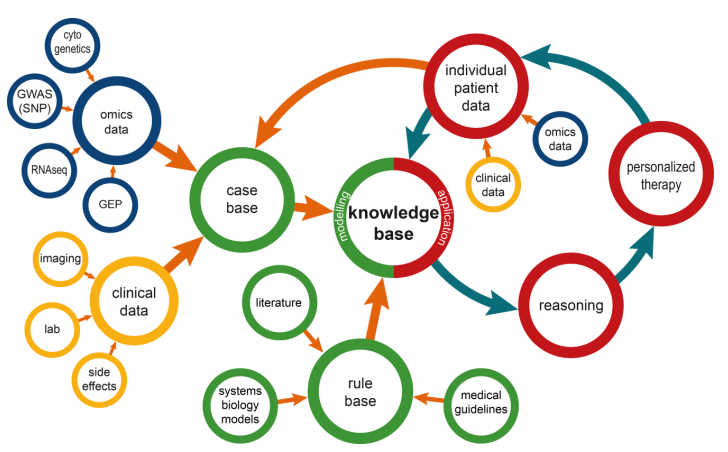
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 18–24:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Dagliati FrontInDigiHum2018 5.jpg|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Dagliati FrontInDigiHum2018 5.jpg|240px]]</div> | ||
'''"[[Journal:Big data as a driver for clinical decision support systems: A learning health systems perspective|Big data as a driver for clinical decision support systems: A learning health systems perspective]]"''' | '''"[[Journal:Big data as a driver for clinical decision support systems: A learning health systems perspective|Big data as a driver for clinical decision support systems: A learning health systems perspective]]"''' | ||
| Line 29: | Line 176: | ||
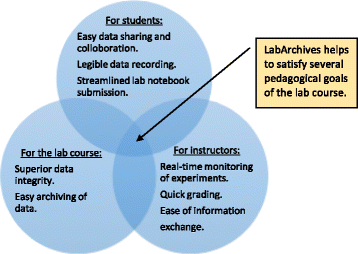
'''"[[Journal:Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory|Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory]]"''' | '''"[[Journal:Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory|Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory]]"''' | ||
[[Electronic laboratory notebook]]s (ELNs) are better equipped than paper [[laboratory notebook]]s (PLNs) to handle present-day life science and engineering experiments that generate large data sets and require high levels of data integrity. But limited training and a lack of workforce with ELN knowledge have restricted the use of ELN in academic and industry research [[Laboratory|laboratories]], which still rely on cumbersome PLNs for record keeping. We used [[LabArchives, LLC|LabArchives]], a cloud-based ELN in our bioprocess engineering lab course to train students in electronic record keeping, good documentation practices (GDPs), and [[data integrity]]. | [[Electronic laboratory notebook]]s (ELNs) are better equipped than paper [[laboratory notebook]]s (PLNs) to handle present-day life science and engineering experiments that generate large data sets and require high levels of data integrity. But limited training and a lack of workforce with ELN knowledge have restricted the use of ELN in academic and industry research [[Laboratory|laboratories]], which still rely on cumbersome PLNs for record keeping. We used [[Vendor:LabArchives, LLC|LabArchives]], a cloud-based ELN in our bioprocess engineering lab course to train students in electronic record keeping, good documentation practices (GDPs), and [[data integrity]]. | ||
Implementation of ELN in the bioprocess engineering lab course, an analysis of user experiences, and our development actions to improve ELN training are presented here. ELN improved pedagogy and learning outcomes of the lab course through streamlined workflow, quick data recording and archiving, and enhanced data sharing and collaboration. It also enabled superior data integrity, simplified information exchange, and allowed real-time and remote monitoring of experiments. ('''[[Journal:Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory|Full article...]]''')<br /> | Implementation of ELN in the bioprocess engineering lab course, an analysis of user experiences, and our development actions to improve ELN training are presented here. ELN improved pedagogy and learning outcomes of the lab course through streamlined workflow, quick data recording and archiving, and enhanced data sharing and collaboration. It also enabled superior data integrity, simplified information exchange, and allowed real-time and remote monitoring of experiments. ('''[[Journal:Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory|Full article...]]''')<br /> | ||
Latest revision as of 20:36, 3 April 2024
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 - 2023 - 2024 |
Featured article of the week archive - 2018
Welcome to the LIMSwiki 2018 archive for the Featured Article of the Week.
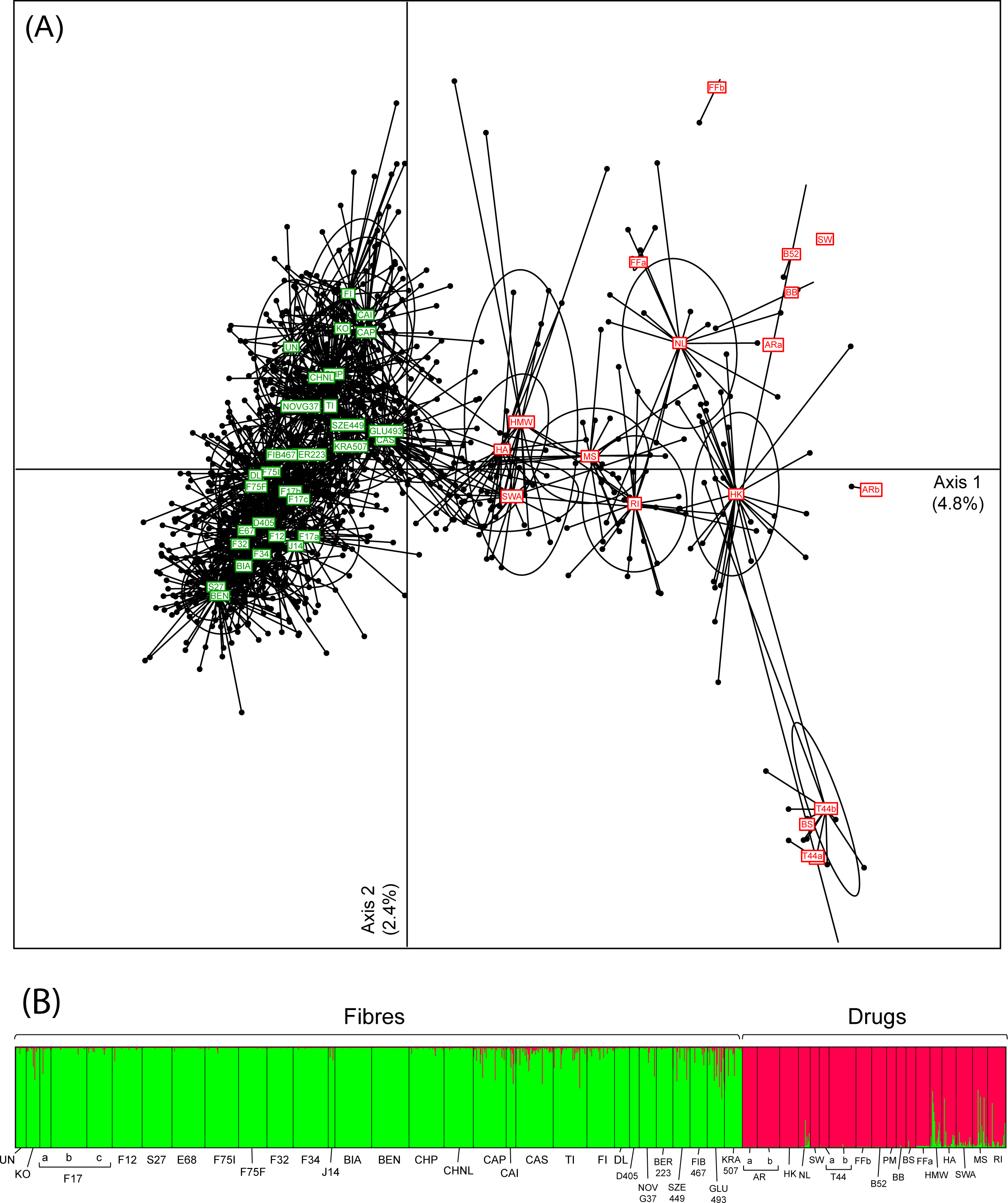
Featured article of the week: December 17–31:"Approaches to medical decision-making based on big clinical data" The paper discusses different approaches to building a clinical decision support system based on big data. The authors sought to abstain from any data reduction and apply universal teaching and big data processing methods independent of disease classification standards. The paper assesses and compares the accuracy of recommendations among three options: case-based reasoning, simple single-layer neural network, and probabilistic neural network. Further, the paper substantiates the assumption regarding the most efficient approach to solving the specified problem. (Full article...)
|