Difference between revisions of "Main Page/Featured article of the week/2019"
Shawndouglas (talk | contribs) (Added last week's article of the week) |
Shawndouglas (talk | contribs) (Ombox header) |
||
| (32 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{ombox | {{ombox | ||
| type = notice | | type = notice | ||
| text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - [[Main Page/Featured article of the week/2017|2017]] - [[Main Page/Featured article of the week/2018|2018]] - 2019 | | text = If you're looking for other "Article of the Week" archives: [[Main Page/Featured article of the week/2014|2014]] - [[Main Page/Featured article of the week/2015|2015]] - [[Main Page/Featured article of the week/2016|2016]] - [[Main Page/Featured article of the week/2017|2017]] - [[Main Page/Featured article of the week/2018|2018]] - 2019 - [[Main Page/Featured article of the week/2020|2020]] - [[Main Page/Featured article of the week/2021|2021]] - [[Main Page/Featured article of the week/2022|2022]] - [[Main Page/Featured article of the week/2023|2023]] - [[Main Page/Featured article of the week/2024|2024]] | ||
}} | }} | ||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 3–9:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 23–31:</h2> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Parker DataSciJourn2019 18-1.png|240px]]</div> | |||
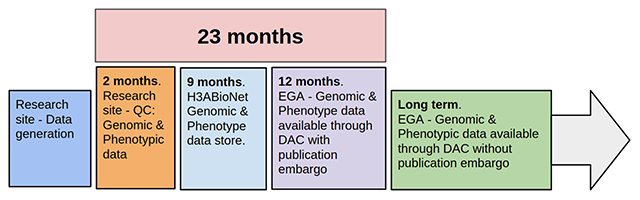
'''"[[Journal:Building infrastructure for African human genomic data management|Building infrastructure for African human genomic data management]]"''' | |||
Human [[Genomics|genomic]] data are large and complex, and require adequate infrastructure for secure storage and transfer. The [[National Institutes of Health]] (NIH) and The Wellcome Trust have funded multiple projects on genomic research, including the Human Heredity and Health in Africa (H3Africa) initiative, and data are required to be deposited into the public domain. The European Genome-phenome Archive (EGA) is a repository for [[Sequencing|sequence]] and genotype data where data access is controlled by access committees. Access is determined by a formal application procedure for the purpose of secure storage and distribution, which must be in line with the informed consent of the study participants. H3Africa researchers based in Africa and generating their own data can benefit tremendously from the data sharing capabilities of the internet by using the appropriate technologies. The H3Africa Data Archive is an effort between the H3Africa data generating projects, H3ABioNet, and the EGA to store and submit genomic data to public repositories. ('''[[Journal:Building infrastructure for African human genomic data management|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 16–22:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Galliano JofPathInfo2019 10.jpg|240px]]</div> | |||
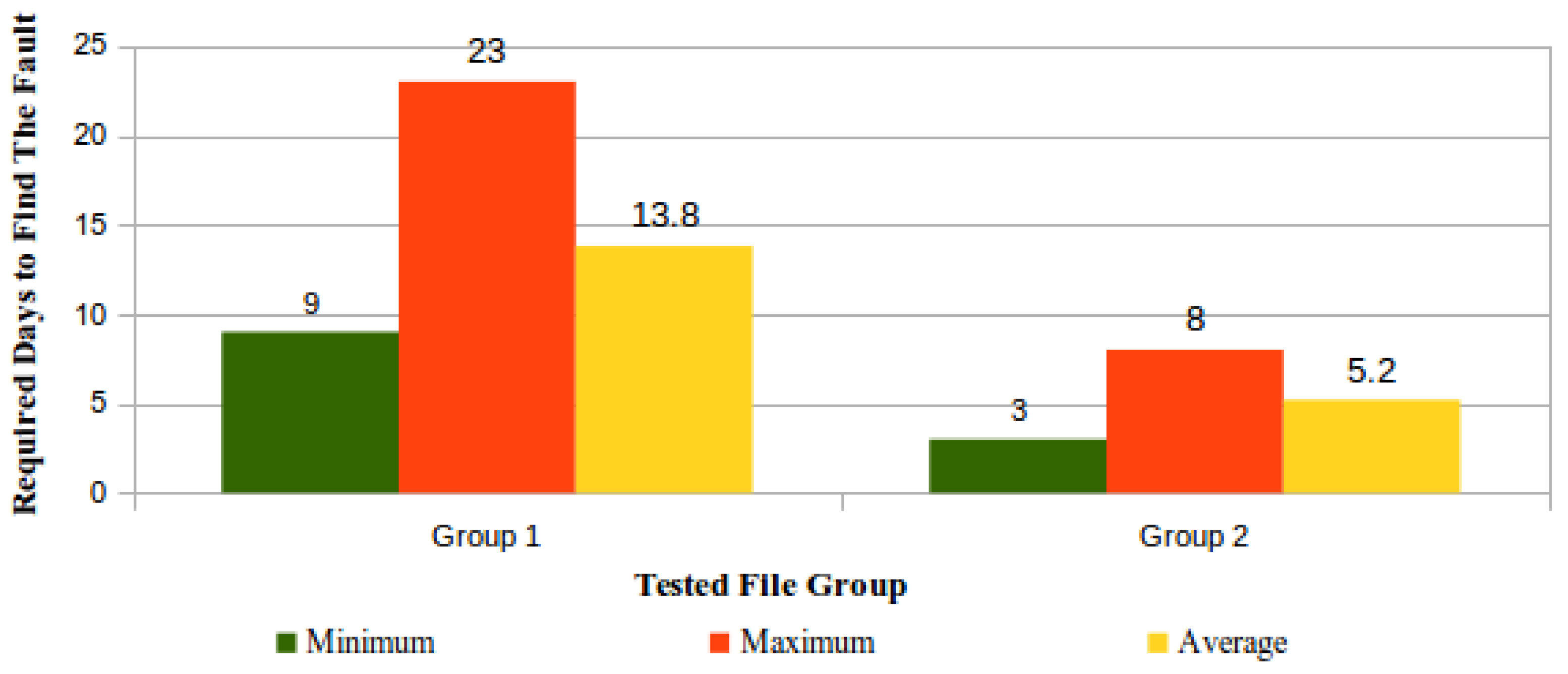
'''"[[Journal:Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory|Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory]]"''' | |||
[[Barcode]]-driven [[workflow]]s reduce patient identification errors. Missing process timestamp data frequently confound our health system's pending lists and appear as actions left undone. Anecdotally, it was noted that missing data could be found when there is procedure noncompliance. This project was developed to determine if missing timestamp data in the histology barcode-driven workflow correlated with other process variations, procedure noncompliance, or is an indicator of workflows needing focus for improvement projects. Data extracts of timestamp data from January 1, 2018 to December 15, 2018 for the major histology process steps were analyzed for missing data. Case-level analysis to determine the presence or absence of expected barcoding events was performed on 1031 surgical pathology cases to determine the cause of the missing data and determine if additional data variations or procedure noncompliance events were present. The data variations were classified according to a scheme defined in the study. ('''[[Journal:Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 9–15:</h2> | |||
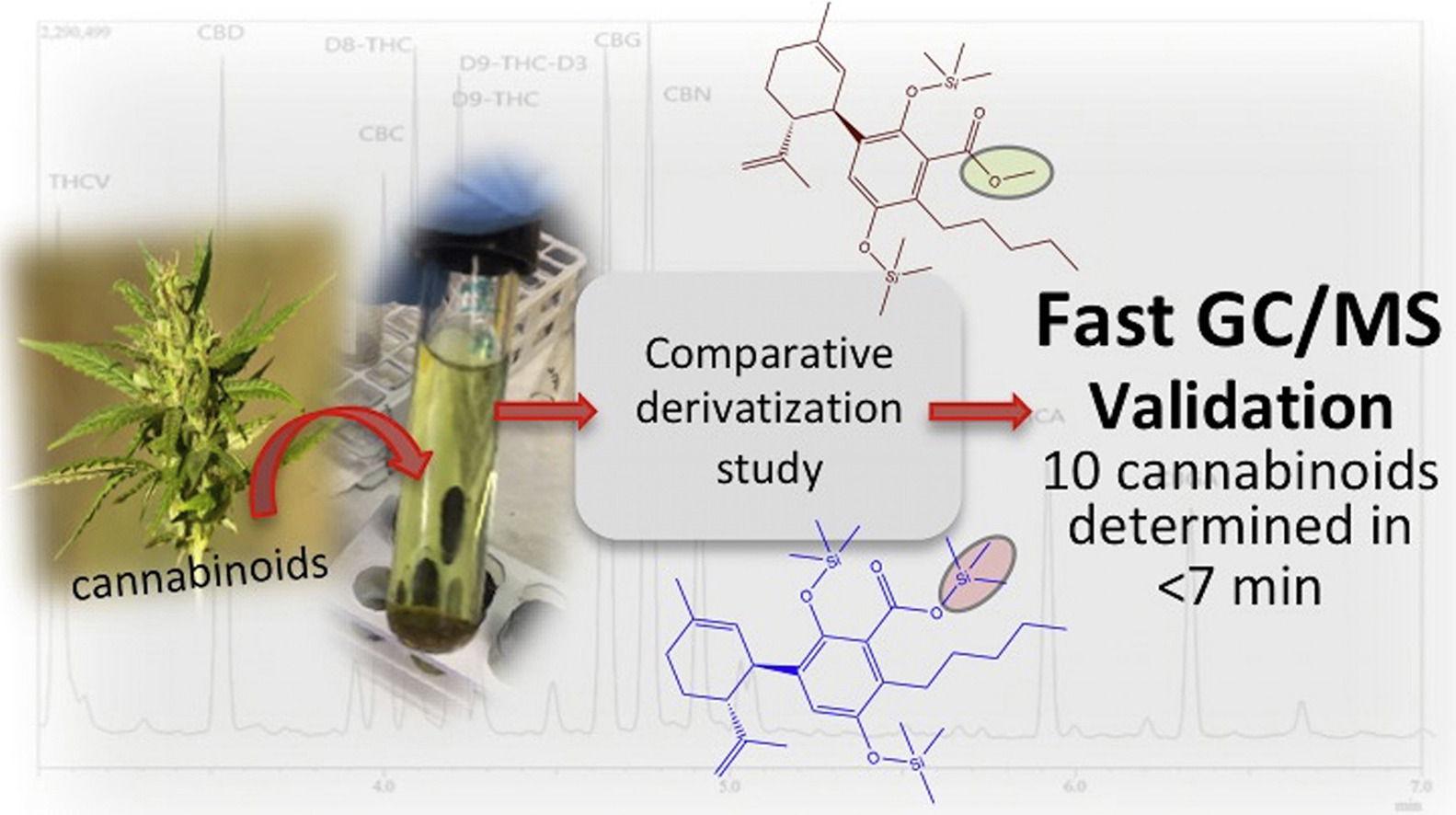
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig0 Cardenia JofFoodDrugAnal2018 26-4.jpg|240px]]</div> | |||
'''"[[Journal:Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L|Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L]]"''' | |||
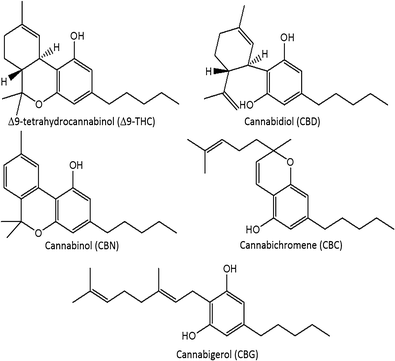
A routine method for determining [[wikipedia:Cannabinoid|cannabinoids]] in ''Cannabis sativa'' L. [[wikipedia:Inflorescence|inflorescence]], based on fast [[gas chromatography]] coupled to [[mass spectrometry]] (fast GC-MS), was developed and validated. To avoid the [[wikipedia:Decarboxylation|decarboxylation]] of the carboxyl group of cannabinoids, different derivatization approaches—i.e., silylation and esterification (diazomethane-mediated) reagents and solvents (pyridine or ethyl acetate)—were tested. The methylation significantly increased the signal-to-noise ratio of all carboxylic cannabinoids, except for cannabigerolic acid (CBGA). Since [[wikipedia:Diazomethane|diazomethane]] is not commercially available, is considered a hazardous reactive, and requires one-day synthesis by specialized chemical staff, the process of silylation was used along the entire validation of a routine method. The method gave a fast (total analysis time < 7.0 min) and satisfactory resolution (R > 1.1), with a good repeatability (intraday < 8.38%; interday < 11.10%) and sensitivity (LOD < 11.20 ng/mL). ('''[[Journal:Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: December 2–8:</h2> | |||
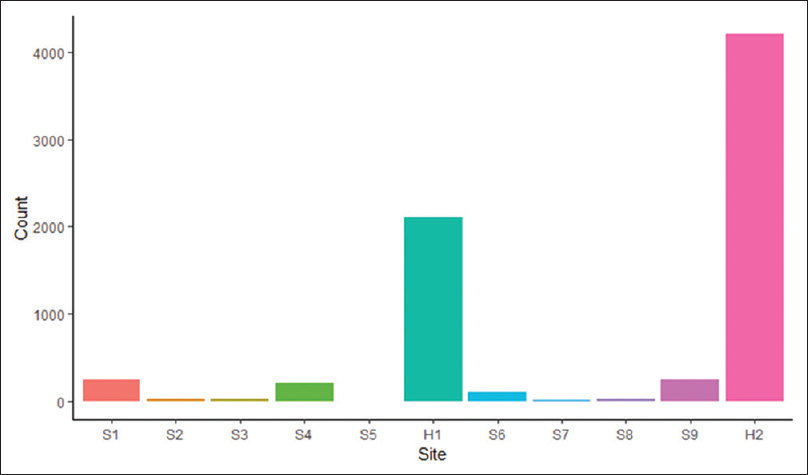
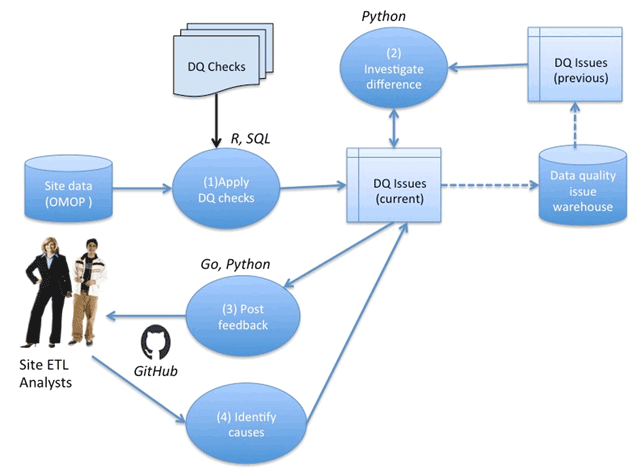
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Khare eGEMs2019 7-1.png|240px]]</div> | |||
'''"[[Journal:Design and refinement of a data quality assessment workflow for a large pediatric research network|Design and refinement of a data quality assessment workflow for a large pediatric research network]]"''' | |||
Clinical data research networks (CDRNs) aggregate [[electronic health record]] (EHR) data from multiple hospitals to enable large-scale research. A critical operation toward building a CDRN is conducting continual evaluations to optimize [[wikipedia:Data quality|data quality]]. The key challenges include determining the assessment coverage on big datasets, handling data variability over time, and facilitating communication with data teams. This study presents the evolution of a systematic workflow for data quality assessment in CDRNs. | |||
Using a specific CDRN as a use case, a workflow was iteratively developed and packaged into a toolkit. The resultant toolkit comprises 685 data quality checks to identify any data quality issues, procedures to reconciliate with a history of known issues, and a contemporary GitHub-based reporting mechanism for organized tracking. | |||
During the first two years of network development, the toolkit assisted in discovering over 800 data characteristics and resolving over 1400 programming errors. Longitudinal analysis indicated that the variability in time to resolution (15day mean, 24day IQR) is due to the underlying cause of the issue, perceived importance of the domain, and the complexity of assessment. ('''[[Journal:Design and refinement of a data quality assessment workflow for a large pediatric research network|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 25–December 1:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Palmieri Molecules2019 24-19.png|240px]]</div> | |||
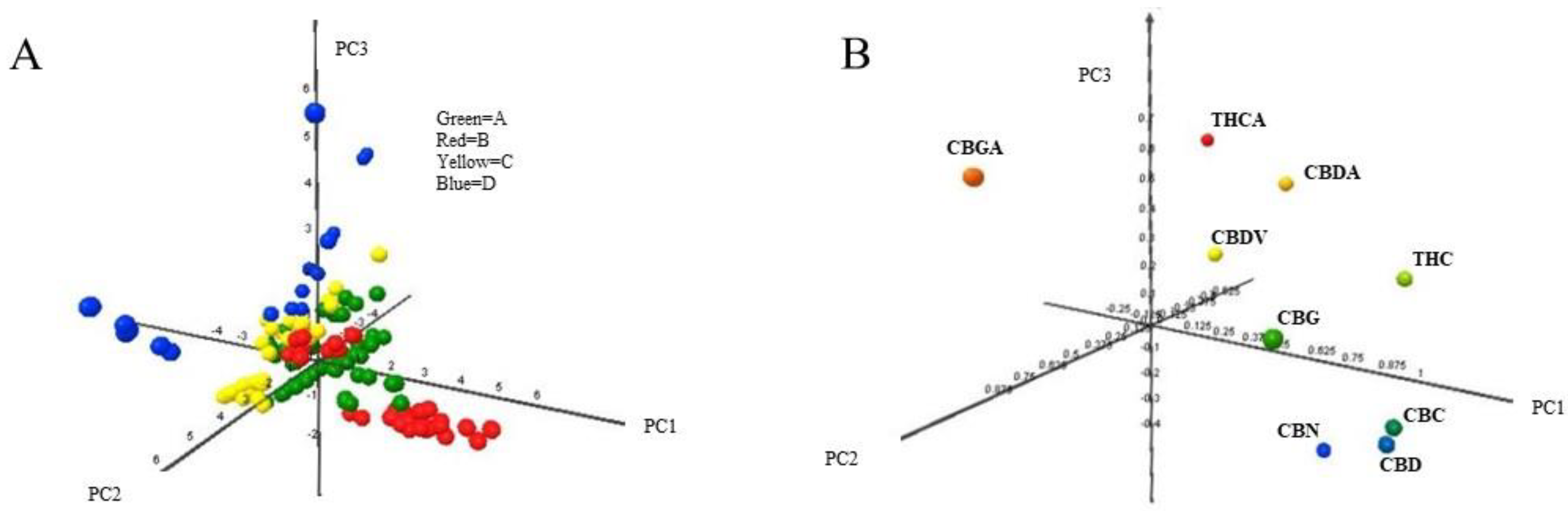
'''"[[Journal:Identification of Cannabis sativa L. (hemp) retailers by means of multivariate analysis of cannabinoids|Identification of Cannabis sativa L. (hemp) retailers by means of multivariate analysis of cannabinoids]]"''' | |||
In this work, the concentration of nine [[wikipedia:Cannabinoid|cannabinoid]]s—six neutral cannabinoids (THC, CBD, CBC, CBG, CBN, and CBDV) and three acidic cannabinoids (THCA, CBGA, and CBDA)—was used to identify the Italian retailers of ''[[wikipedia:Cannabis sativa|Cannabis sativa]]'' L. ([[wikipedia:Hemp|hemp]]), reinforcing the idea that the practice of categorizing hemp samples only using THC and CBD is inadequate. A [[high-performance liquid chromatography]]–[[tandem mass spectrometry]] (HPLC-MS/MS) method was developed for screening and simultaneously analyzing the nine cannabinoids in 161 hemp samples sold by four retailers located in different Italian cities. The hemp samples dataset was analyzed by [[wikipedia:Univariate analysis|univariate]] and [[wikipedia:Multivariate analysis|multivariate analysis]], with the aim to identify the associated hemp retailers without using any other [[information]] on the hemp samples such as [[wikipedia:Cannabis strains|''Cannabis'' strains]], seeds, soil and cultivation characteristics, geographical origin, product storage, etc. The univariate analysis highlighted that the hemp samples could not be differentiated by using any of the nine cannabinoids analyzed. ('''[[Journal:Identification of Cannabis sativa L. (hemp) retailers by means of multivariate analysis of cannabinoids|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 18–24:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Poirier DataScienceJournal2019 18-1.png|240px]]</div> | |||
'''"[[Journal:Data sharing at scale: A heuristic for affirming data cultures|Data sharing at scale: A heuristic for affirming data cultures]]"''' | |||
Addressing the most pressing contemporary social, environmental, and technological challenges will require integrating insights and sharing data across disciplines, geographies, and cultures. Strengthening international data sharing networks will not only demand advancing technical, legal, and logistical infrastructure for publishing data in open, accessible formats; it will also require recognizing, respecting, and learning to work across diverse data cultures. This essay introduces a heuristic for pursuing richer characterizations of the “data cultures” at play in international, interdisciplinary data sharing. The heuristic prompts cultural analysts to query the contexts of data sharing for a particular discipline, institution, geography, or project at seven scales: the meta, macro, meso, micro, techno, data, and nano. The essay articulates examples of the diverse cultural forces acting upon and interacting with researchers in different communities at each scale. The heuristic we introduce in this essay aims to elicit from researchers the beliefs, values, practices, incentives, and restrictions that impact how they think about and approach data sharing. Rather than represent an effort to iron out differences between disciplines, this essay instead intends to showcase and affirm the diversity of traditions and modes of analysis that have shaped how data gets collected, organized, and interpreted in diverse settings. ('''[[Journal:Data sharing at scale: A heuristic for affirming data cultures|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 11–17:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Wang BMCMedInfoDecMak2019 19-1.png|240px]]</div> | |||
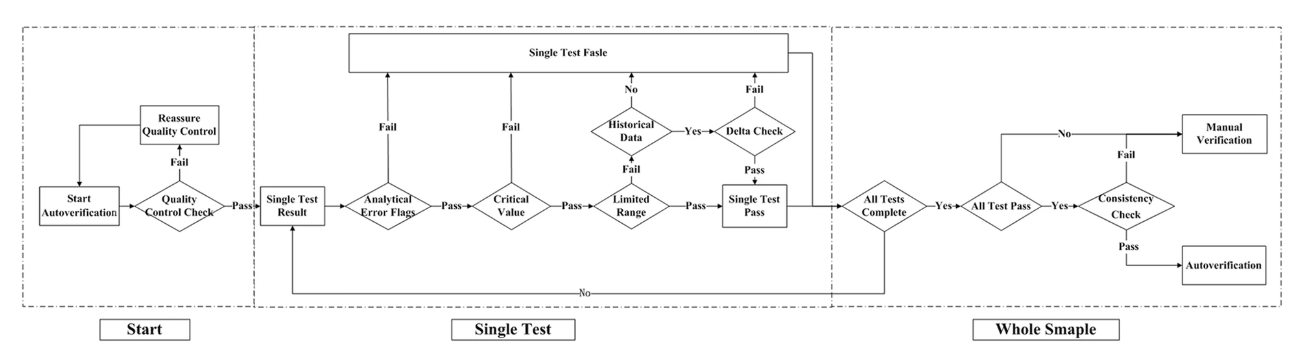
'''"[[Journal:Design and evaluation of a LIS-based autoverification system for coagulation assays in a core clinical laboratory|Design and evaluation of a LIS-based autoverification system for coagulation assays in a core clinical laboratory]]"''' | |||
n autoverification system for coagulation consists of a series of rules that allows normal data to be released without manual verification. With new advances in [[medical informatics]], the [[laboratory information system]] (LIS) has growing potential for the use of autoverification, allowing rapid and accurate verification of [[clinical laboratory]] tests. The purpose of the study is to develop and evaluate a LIS-based autoverification system for validation and efficiency. | |||
Autoverification decision rules—including quality control, analytical error flag, critical value, limited range check, delta check, and logical check rules, as well as patient’s historical information—were integrated into the LIS. Autoverification limit ranges was constructed based on 5% and 95% percentiles. The four most commonly used coagulation assays—prothrombin time (PT), activated partial thromboplastin time (APTT), thrombin time (TT), and fibrinogen (FBG)—were followed by the autoverification protocols. ('''[[Journal:Design and evaluation of a LIS-based autoverification system for coagulation assays in a core clinical laboratory|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: November 4–10:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Hodhod IntJofOnlineBiomedEng2019 15-3.png|240px]]</div> | |||
'''"[[Journal:CyberMaster: An expert system to guide the development of cybersecurity curricula|CyberMaster: An expert system to guide the development of cybersecurity curricula]]"''' | |||
The growing number of reported cyberattacks poses a difficult challenge to individuals, governments, and organizations. Adequate protection of [[information]] systems urgently requires a [[cybersecurity]]-educated workforce trained using a curriculum that covers the essential skills required for different cybersecurity work roles. The goal of the CyberMaster [[expert system]] is to assist inexperienced instructors with cybersecurity course design. It is an intelligent system that uses visual feedback to guide the user through the design process. Initial test executions show the promise of such a system in addressing the enormous shortage of cybersecurity experts currently available for designing courses and training programs. ('''[[Journal:CyberMaster: An expert system to guide the development of cybersecurity curricula|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 21–November 3:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab2 Valdes-Donoso CaliforniaAg2019 73-3.jpg|240px]]</div> | |||
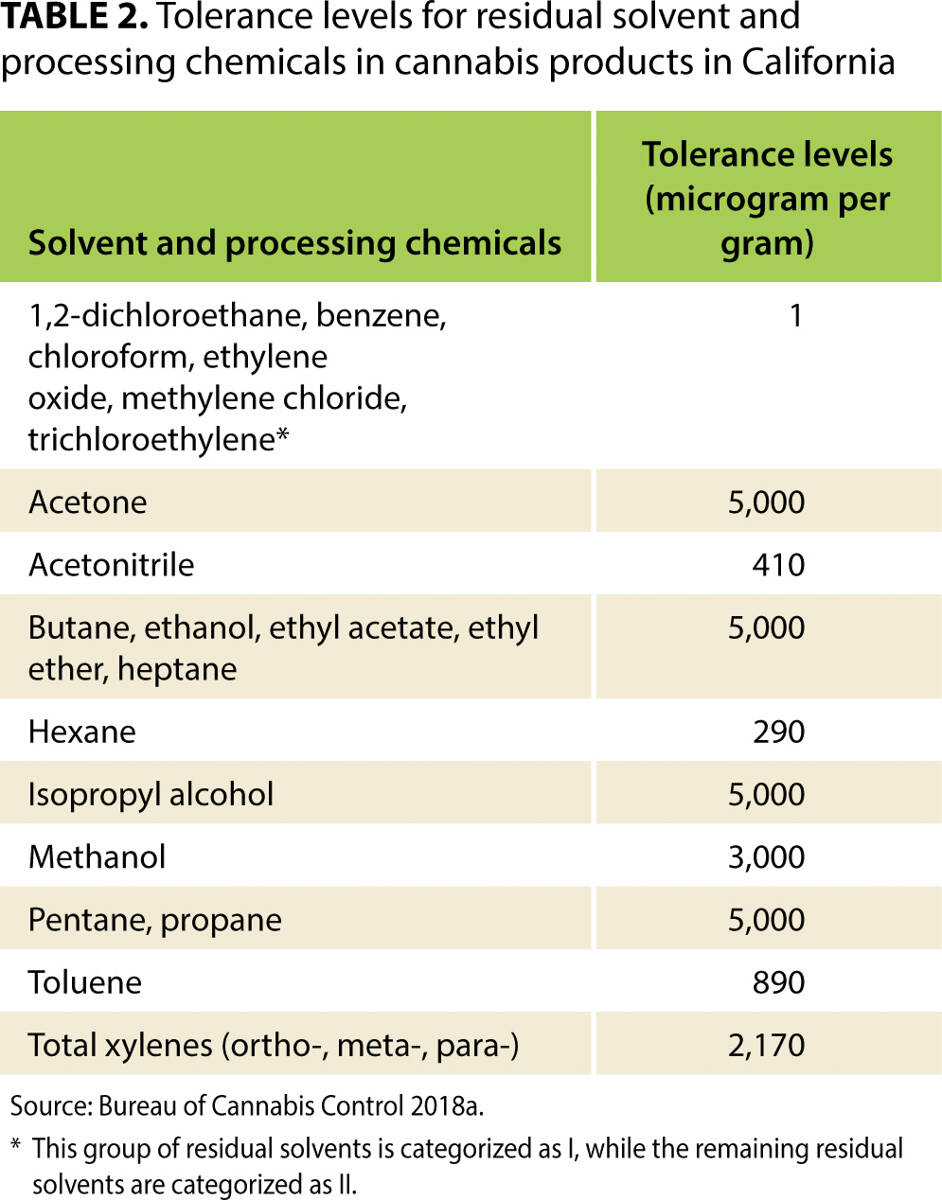
'''"[[Journal:Costs of mandatory cannabis testing in California|Costs of mandatory cannabis testing in California]]"''' | |||
Every batch of [[wikipedia:Cannabis|cannabis]] sold legally in California must be tested for more than 100 contaminants. These contaminants include 66 pesticides, for 21 of which the state's tolerance is zero. For many other substances, tolerance levels are much lower than those allowed for food products in California. This article reviews the state's testing [[Regulatory compliance|regulations]] in context—including maximum allowable tolerance levels—and uses primary data collected from California's major cannabis testing [[Laboratory|laboratories]] and several cannabis testing equipment manufacturers, as well as a variety of expert opinions, to estimate the cost per pound of testing under the state's framework. We also estimate the cost of collecting [[Sample (material)|samples]], which depends on the distance between cannabis distributors and laboratories. We find that, if a batch fails mandatory tests, the value of cannabis that must be destroyed accounts for a large share of total testing costs, more than the cost of the tests that laboratories perform. Findings from this article will help readers understand the effects of California's testing regime on the price of legal cannabis in the state, and understand how testing may add value to products that have passed a series of tests that aim to validate their safety. ('''[[Journal:Costs of mandatory cannabis testing in California|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 14–20:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Armstrong FrontMarineSci2019 6.jpg|240px]]</div> | |||
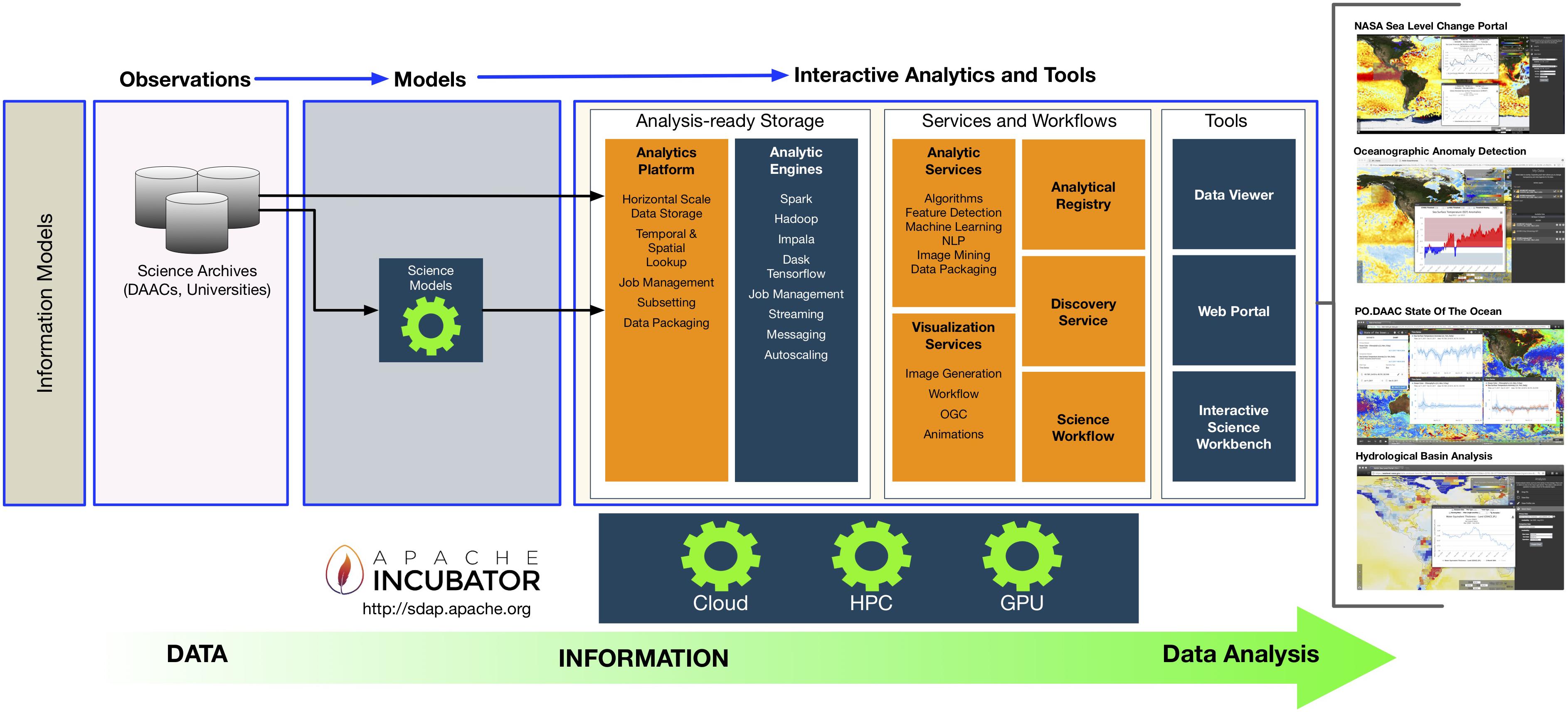
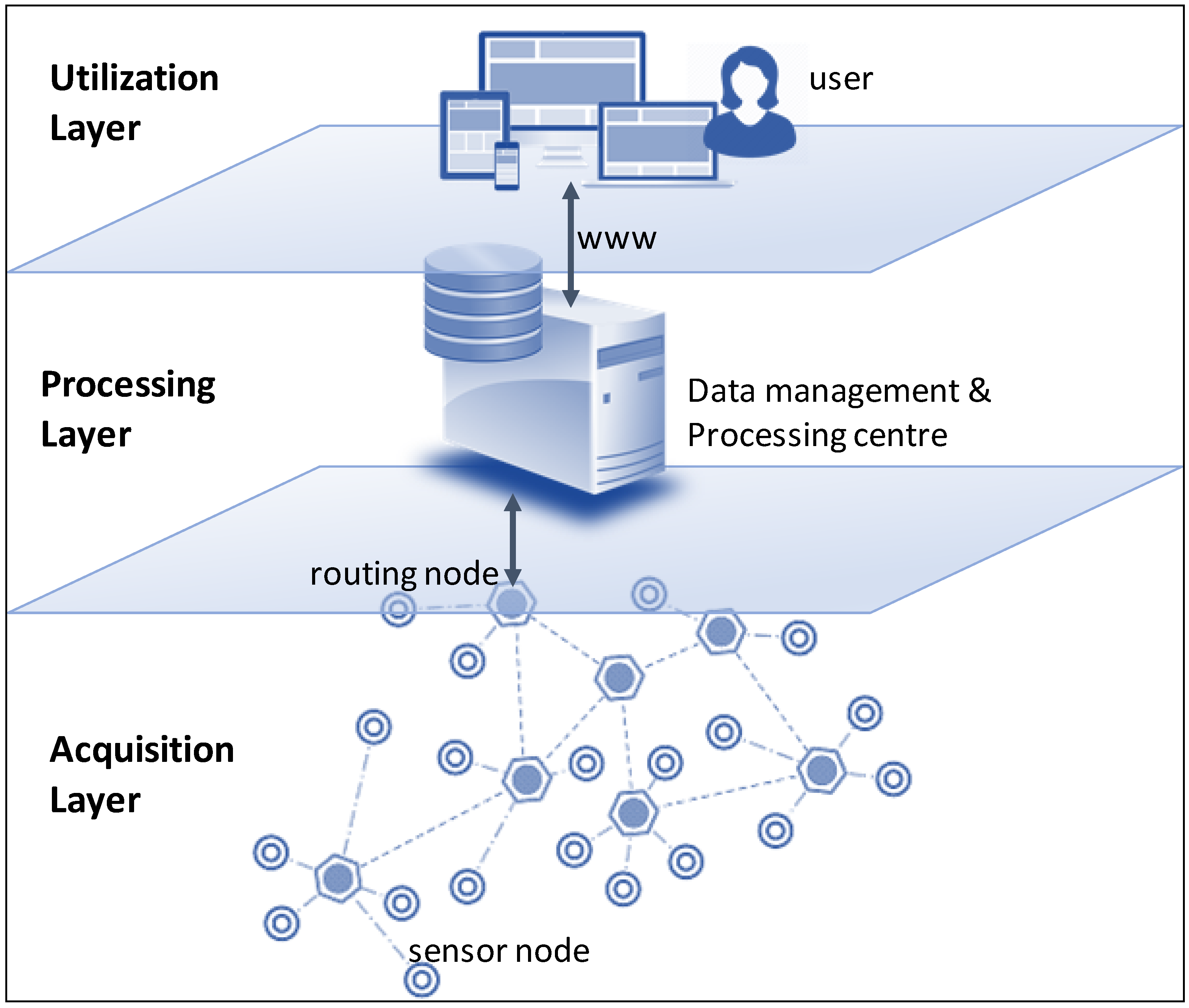
'''"[[Journal:An integrated data analytics platform|An integrated data analytics platform]]"''' | |||
A scientific integrated data analytics platform (IDAP) is an environment that enables the confluence of resources for scientific investigation. It harmonizes data, tools, and computational resources to enable the research community to focus on the investigation rather than spending time on security, data preparation, management, etc. OceanWorks is a National Aeronautics and Space Administration (NASA) technology integration project to establish a [[Cloud computing|cloud-based]] integrated ocean science [[Data analysis|data analytics]] platform for managing ocean science research data at NASA’s Physical Oceanography Distributed Active Archive Center (PO.DAAC). The platform focuses on advancement and maturity by bringing together several NASA open-source, big data projects for parallel analytics, anomaly detection, ''in situ''-to-satellite data matching, quality-screened data subsetting, search relevancy, and data discovery. ('''[[Journal:An integrated data analytics platform|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: October 7–13:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Annane IntJInterMobileTech2019 13-4.png|240px]]</div> | |||
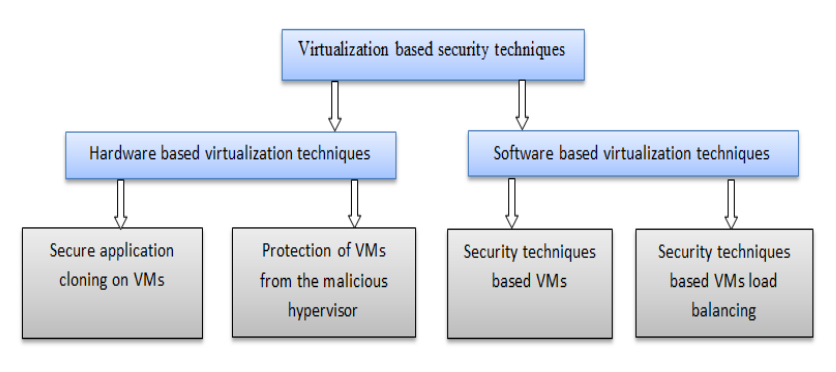
'''"[[Journal:Virtualization-based security techniques on mobile cloud computing: Research gaps and challenges|Virtualization-based security techniques on mobile cloud computing: Research gaps and challenges]]"''' | |||
The principle constraints of mobile devices are their limited resources, including processing capability, storage space, and battery life. However, [[cloud computing]] offers a means of vast computing resources and services. With it a new idea emerged, the inclusion of cloud computing into mobile devices such as smartphones, tablet, and other personal digital assistants (PDA) to augment their capacities, providing a robust technology called mobile cloud computing (MCC). Although MCC has brought many advantages to mobile users, it also still suffers from the security and privacy issues of data while hosted on virtual machines (VM) on remote cloud’s servers. Currently, the eyes of security experts are turned towards the virtualization-based security techniques used either on the cloud or on mobile devices. The new challenge is to develop secure methods in order to authenticate highly sensitive digital content. ('''[[Journal:Virtualization-based security techniques on mobile cloud computing: Research gaps and challenges|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 30–October 6:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Greene DataScienceJ2019 18-1.jpg|240px]]</div> | |||
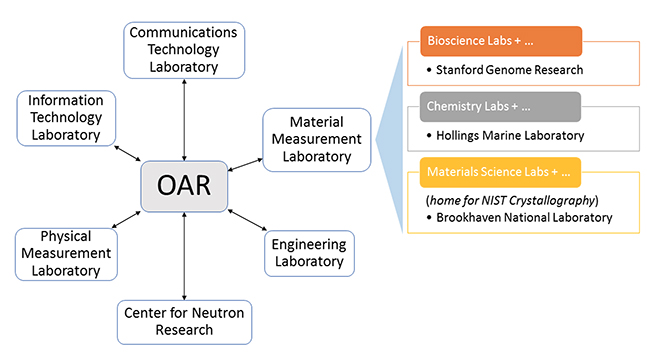
'''"[[Journal:Building open access to research (OAR) data infrastructure at NIST|Building open access to research (OAR) data infrastructure at NIST]]"''' | |||
As a National Metrology Institute (NMI), the U.S. National Institute of Standards and Technology (NIST) scientists, engineers, and technology experts conduct research across a full spectrum of physical science domains. NIST is a non-regulatory agency within the U.S. Department of Commerce with a mission to promote U.S. innovation and industrial competitiveness by advancing measurement science, standards, and technology in ways that enhance economic security and improve our quality of life. NIST research results in the production and distribution of standard [[Reference laboratory#Reference measurement and calibration|reference materials]], [[[[Reference laboratory#Reference measurement and calibration|calibration services]], and datasets. These are generated from a wide range of complex [[laboratory]] instrumentation, expert analyses, and calibration processes. In response to a government open data policy, and in collaboration with the broader research community, NIST has developed a federated Open Access to Research (OAR) scientific data infrastructure aligned with FAIR (findable, accessible, interoperable, reusable) data principles. ('''[[Journal:Building open access to research (OAR) data infrastructure at NIST|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 23–29:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Baratta FrontPharmaco2019 10.jpg|240px]]</div> | |||
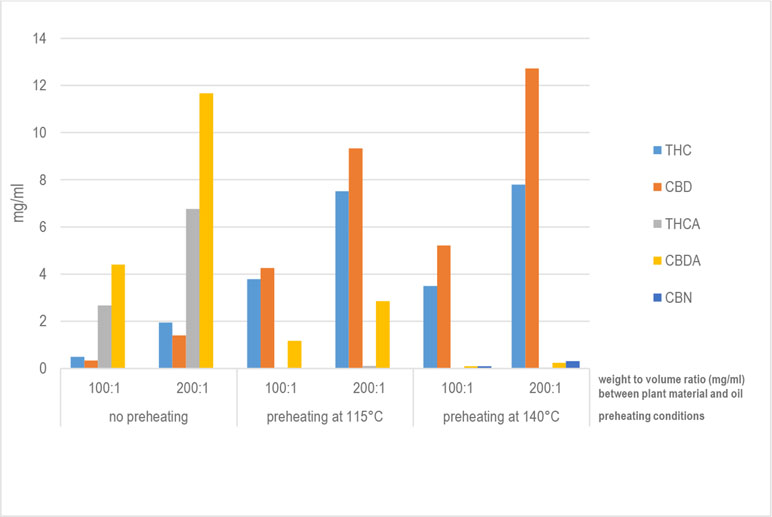
'''"[[Journal:Development of standard operating protocols for the optimization of Cannabis-based formulations for medical purposes|Development of standard operating protocols for the optimization of Cannabis-based formulations for medical purposes]]"''' | |||
Under current legislation in Italy, using the ''[[wikipedia:Cannabis|Cannabis]]'' plant for medical purposes requires administering it orally in the form of a decoction or as ''Cannabis'' oil extract. The scientific literature reports a number of preparation methods, mainly for oils, but no study is available that compares thoroughly, from a technological viewpoint, the ''Cannabis''-based formulations currently administered to patients. With this in mind, this research work aimed to carry out specific formulation studies to design standard operating procedures for the preparation and optimization of ''Cannabis''-based galenic formulations. Both decoctions and oils were prepared under different operating conditions to identify the most efficient process for the production of formulations with a high concentration of [[wikipedia:Decarboxylation|decarboxylated]] [[wikipedia:Tetrahydrocannabinol|delta-9-tetrahydrocannabinol]] (THC) and [[wikipedia:Cannabidiol|cannabidiol]] (CBD). Regarding ''Cannabis'' oil, a new procedure has been developed that allows significantly higher recovery rates for THC and CBD compared with those for water-based extraction methods (decoction) and those for oil-based methods currently in use. ('''[[Journal:Development of standard operating protocols for the optimization of Cannabis-based formulations for medical purposes|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 16–22:</h2> | |||
'''"[[Journal:Next steps for access to safe, secure DNA synthesis|Next steps for access to safe, secure DNA synthesis]]"''' | |||
The [[DNA synthesis]] industry has, since the invention of gene-length synthesis, worked proactively to ensure synthesis is carried out securely and safely. Informed by guidance from the U.S. government, several of these companies have collaborated over the last decade to produce a set of best practices for customer and sequence screening prior to manufacture. Taken together, these practices ensure that synthetic DNA is used to advance research that is designed and intended for public benefit. With increasing scale in the industry and expanding capability in the synthetic biology toolset, it is worth revisiting current practices to evaluate additional measures to ensure the continued safety and wide availability of DNA synthesis. Here we encourage specific steps, in part derived from successes in the [[cybersecurity]] community, that can ensure synthesis screening systems stay well ahead of emerging challenges, to continue to enable responsible research advances. [[Artificial gene synthesis|Gene synthesis]] companies, science and technology funders, policymakers, and the scientific community as a whole have a shared duty to continue to minimize risk and maximize the safety and security of DNA synthesis to further power world-changing developments in advanced biological manufacturing, agriculture, drug development, healthcare, and energy. ('''[[Journal:Next steps for access to safe, secure DNA synthesis|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 9–15:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Oyashi GeospatialHlth2019 14-1.png|240px]]</div> | |||
'''"[[Journal:Smart grids and ethics: A case study|Smart grids and ethics: A case study]]"''' | |||
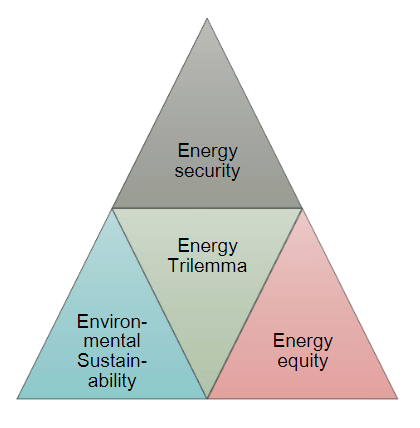
This case study explores the principal ethical issues that occur in the use of smart information systems (SIS) in smart grids and offers suggestions as to how they might be addressed. Key issues highlighted in the literature are reviewed. The empirical case study describes one of the largest distribution system operators (DSOs) in the Netherlands. The aim of this case study is to identify which ethical issues arise from the use of SIS in smart grids, the current efforts of the organization to address them, and whether practitioners are facing additional issues not addressed in current literature. The literature review highlights mainly ethical issues around health and safety, privacy and informed consent, cyber-risks and energy security, affordability, equity, and sustainability. The key topics raised by interviewees revolved around privacy and to some extent [[cybersecurity]]. ('''[[Journal:Smart grids and ethics: A case study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: September 2–8:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Hatzakis ORBITJ2019 2-2.png|240px]]</div> | |||
'''"[[Journal:Japan Aerospace Exploration Agency’s public-health monitoring and analysis platform: A satellite-derived environmental information system supporting epidemiological study|Japan Aerospace Exploration Agency’s public-health monitoring and analysis platform: A satellite-derived environmental information system supporting epidemiological study]]"''' | |||
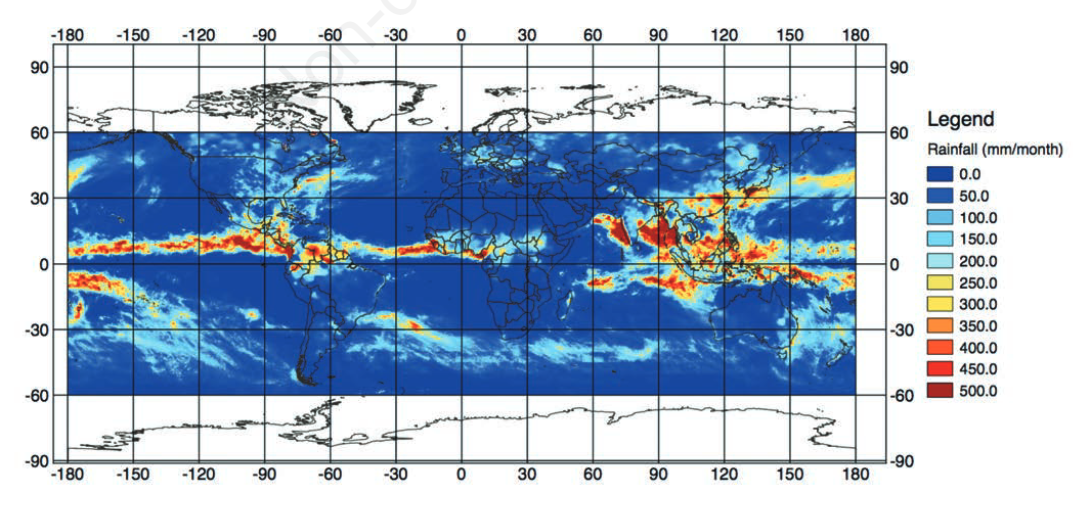
Since the 1970s, Earth-observing satellites collect increasingly detailed [[Environmental monitoring|environmental information]] on land cover, meteorological conditions, environmental variables, and air pollutants. This [[information]] spans the entire globe, and its acquisition plays an important role in epidemiological analysis when ''in situ'' data are unavailable or spatially and/or temporally sparse. In this paper, we present the development of the Japan Aerospace Exploration Agency’s (JAXA) Public-health Monitoring and Analysis Platform, a user-friendly, web-based system providing environmental data on shortwave radiation, rainfall, soil moisture, the normalized difference vegetation index, aerosol optical thickness, land surface temperature and altitude. ('''[[Journal:Japan Aerospace Exploration Agency’s public-health monitoring and analysis platform: A satellite-derived environmental information system supporting epidemiological study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 26–September 1:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Anwar PLOSMed2019 16-5.png|240px]]</div> | |||
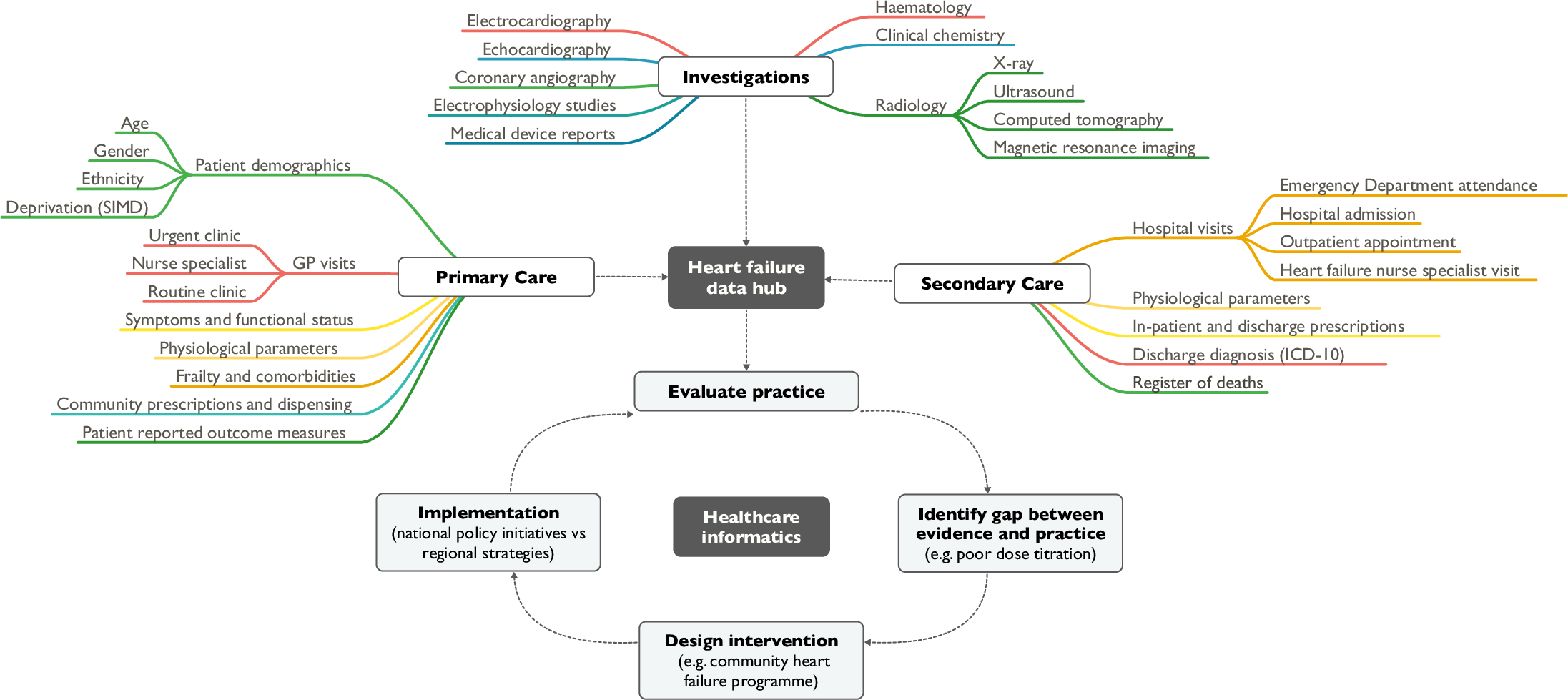
'''"[[Journal:Heart failure and healthcare informatics|Heart failure and healthcare informatics]]"''' | |||
As biomedical research expands our armory of effective, evidence-based therapies, there is a corresponding need for high-quality implementation science—the study of strategies to integrate and embed research advances into [[Health care|clinical practice]]. Large-scale collection and analysis of routinely collected healthcare data may facilitate this in three main ways. Firstly, evaluation of key healthcare metrics can help to identify the areas of practice that differ most from guideline recommendations. Secondly, with sufficiently granular data, it may be possible to detect the underlying drivers of deficiencies in practice. Thirdly, longitudinal data collection should enable us to evaluate large-scale policy initiatives and compare the effectiveness of differing strategies on process and patient outcomes. ('''[[Journal:Heart failure and healthcare informatics|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 19–25:</h2> | |||
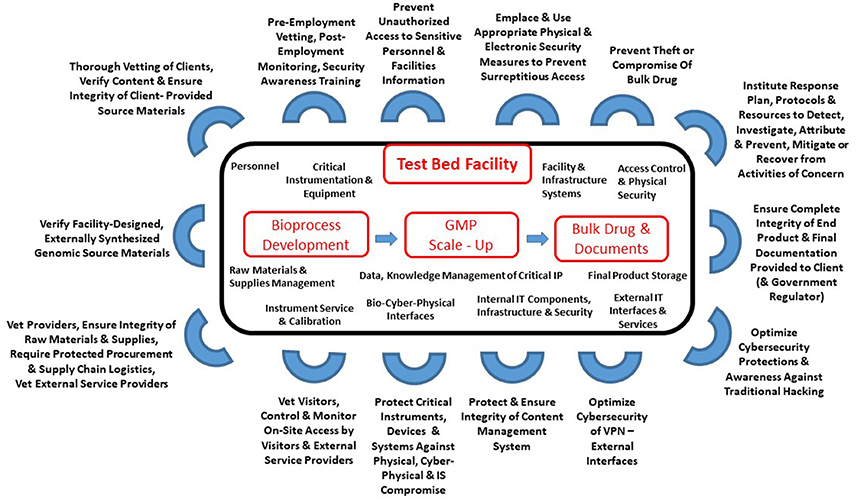
'''"[[Journal:Cyberbiosecurity for biopharmaceutical products|Cyberbiosecurity for biopharmaceutical products]]"''' | |||
Cyberbiosecurity is an emerging discipline that addresses the unique vulnerabilities and threats that occur at the intersection of cyberspace and [[biotechnology]]. Advances in technology and manufacturing are increasing the relevance of cyberbiosecurity to the biopharmaceutical manufacturing community in the United States. Threats may be associated with the biopharmaceutical product itself or with the digital thread of manufacturing of biopharmaceuticals, including those that relate to supply chain and cyberphysical systems. Here, we offer an initial examination of these cyberbiosecurity threats as they stand today, as well as introductory steps toward paths for mitigation of cyberbiosecurity risk for a safer, more secure future. ('''[[Journal:Cyberbiosecurity for biopharmaceutical products|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 12–18:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Matielo Publications2018 6-4.png|240px]]</div> | |||
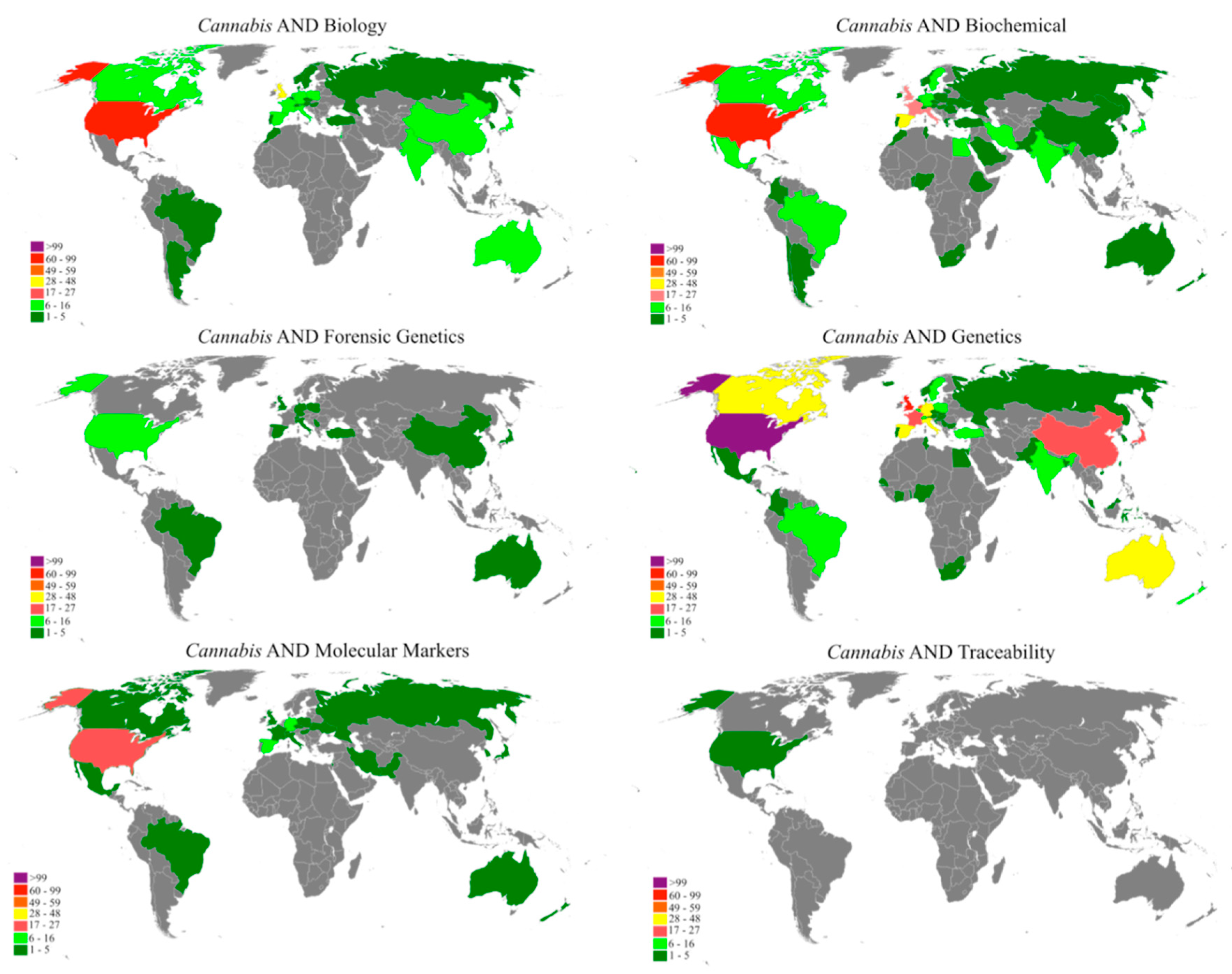
'''"[[Journal:A bibliometric analysis of Cannabis publications: Six decades of research and a gap on studies with the plant|A bibliometric analysis of ''Cannabis'' publications: Six decades of research and a gap on studies with the plant]]"''' | |||
In this study we performed a bibliometric analysis focusing on the general patterns of scientific publications about ''[[wikipedia:Cannabis|Cannabis]]'', revealing their trends and limitations. Publications related to ''Cannabis'', released from 1960 to 2017, were retrieved from the Scopus database using six search terms. The search term “[[wikipedia:Genetics|Genetics]]” returned 53.4% of publications, while “forensic genetics” and “[[wikipedia:Traceability|traceability]]” represented 2.3% and 0.1% of the publications, respectively. However, 43.1% of the studies were not directly related to ''Cannabis'' and, in some cases, ''Cannabis'' was just used as an example in the text. A significant increase in publications was observed after 2001, with most of the publications coming from Europe, followed by North America. Although the term "''Cannabis''" was found in the title, abstract, or keywords of 1284 publications, we detected a historical gap in studies on the plant. ('''[[Journal:A bibliometric analysis of Cannabis publications: Six decades of research and a gap on studies with the plant|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: August 5–11:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Mudge AnalBioChem2017 409-12.gif|240px]]</div> | |||
'''"[[Journal:Leaner and greener analysis of cannabinoids|Leaner and greener analysis of cannabinoids]]"''' | |||
There is an explosion in the number of [[Laboratory|labs]] analyzing [[wikipedia:Cannabinoid|cannabinoids]] in marijuana ([[wikipedia:Cannabis|''Cannabis sativa'' L.]], Cannabaceae); however, existing methods are inefficient, require expert analysts, and use large volumes of potentially environmentally damaging [[wikipedia:Solvent|solvents]]. The objective of this work was to develop and validate an accurate method for analyzing cannabinoids in cannabis raw materials and finished products that is more efficient and uses fewer toxic solvents. A method using [[high-performance liquid chromatography]] (HPLC) with [[Chromatography detector|diode-array detection]] (DAD) was developed for eight cannabinoids in ''Cannabis'' flowers and oils using a statistically guided optimization plan based on the principles of green chemistry. A single-laboratory validation determined the linearity, selectivity, accuracy, repeatability, intermediate precision, limit of detection, and limit of quantitation of the method. Amounts of individual cannabinoids above the limit of quantitation in the flowers ranged from 0.02 to 14.9% concentration (w/w), with repeatability ranging from 0.78 to 10.08% relative standard deviation. ('''[[Journal:Leaner and greener analysis of cannabinoids|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 29–August 4:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Brusniak BMCBioinformatics2019 20.png|240px]]</div> | |||
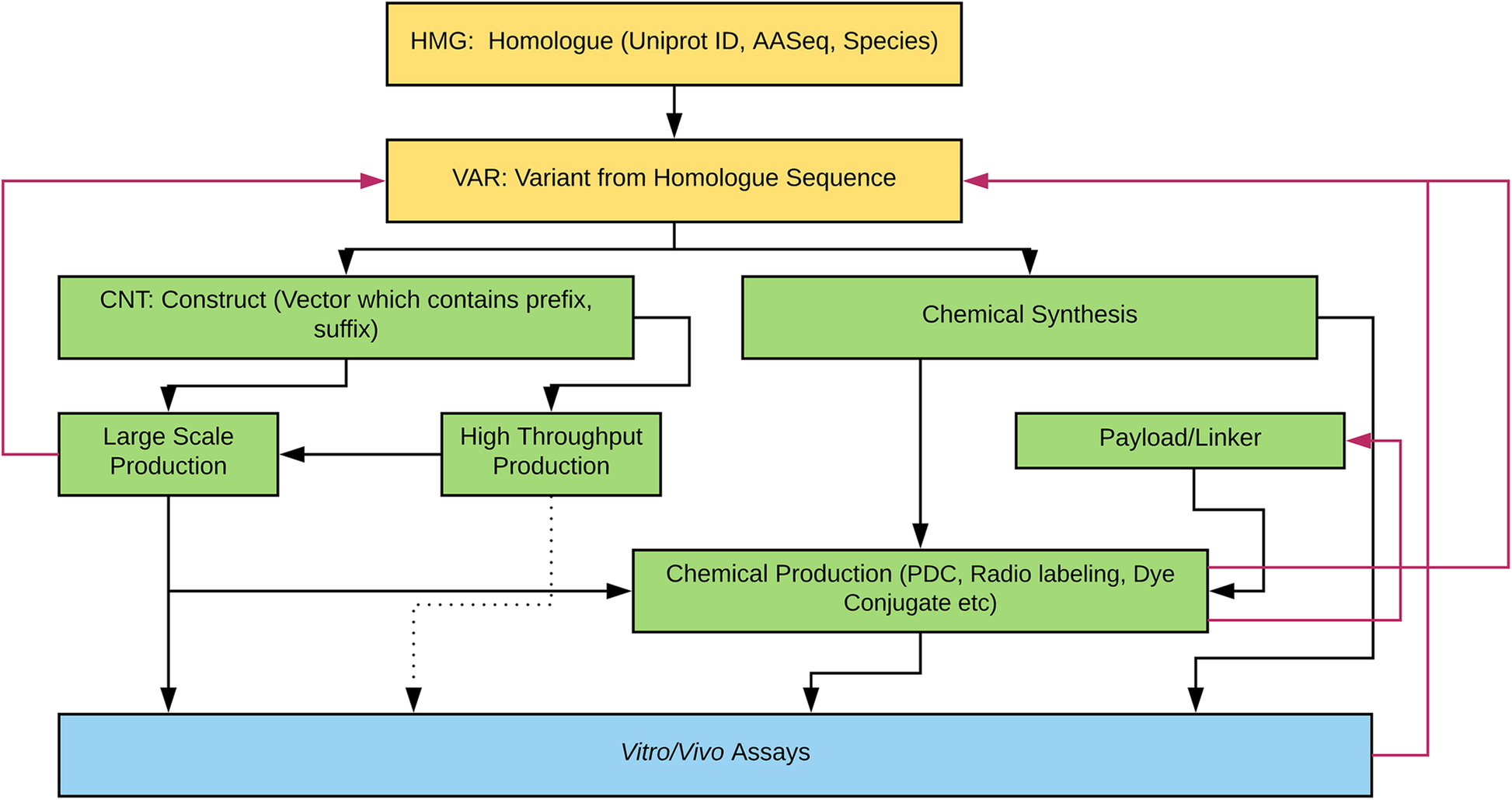
'''"[[Journal:Laboratory information management software for engineered mini-protein therapeutic workflow|Laboratory information management software for engineered mini-protein therapeutic workflow]]"''' | |||
Protein-based therapeutics are one of the fastest growing classes of novel medical interventions in areas such as cancer, infectious disease, and inflammation. Protein engineering plays an important role in the optimization of desired therapeutic properties such as reducing immunogenicity, increasing stability for storage, increasing target specificity, etc. One category of protein therapeutics is nature-inspired bioengineered cystine-dense peptides (CDPs) for various biological targets. These engineered proteins are often further modified by synthetic chemistry. For example, candidate mini-proteins can be conjugated into active small molecule drugs. We refer to modified mini-proteins as "optides" (optimized peptides). To efficiently serve the multidisciplinary lab scientists with varied therapeutic portfolio research goals in a non-commercial setting, a cost-effective, extendable [[laboratory information management system]] (LIMS) is/was needed. ('''[[Journal:Laboratory information management software for engineered mini-protein therapeutic workflow|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 22–28:</h2> | |||
'''"[[Journal:Defending our public biological databases as a global critical infrastructure|Defending our public biological databases as a global critical infrastructure]]"''' | |||
Progress in modern biology is being driven, in part, by the large amounts of freely available data in public resources such as the International Nucleotide Sequence Database Collaboration (INSDC), the world's primary database of biological sequence (and related) [[information]]. INSDC and similar databases have dramatically increased the pace of fundamental biological discovery and enabled a host of innovative therapeutic, diagnostic, and forensic applications. However, as high-value, openly shared resources with a high degree of assumed trust, these repositories share compelling similarities to the early days of the internet. Consequently, as public biological databases continue to increase in size and importance, we expect that they will face the same threats as undefended cyberspace. There is a unique opportunity, before a significant breach and loss of trust occurs, to ensure they evolve with quality and security as a design philosophy rather than costly “retrofitted” mitigations. This perspective article surveys some potential quality assurance and security weaknesses in existing open [[Genomics|genomic]] and [[Proteomics|proteomic]] repositories, describes methods to mitigate the likelihood of both intentional and unintentional errors, and offers recommendations for risk mitigation based on lessons learned from [[cybersecurity]]. ('''[[Journal:Defending our public biological databases as a global critical infrastructure|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 15–21:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig4 Ebnehoseini OAccessMacJofMedSci2019 7-9.png|240px]]</div> | |||
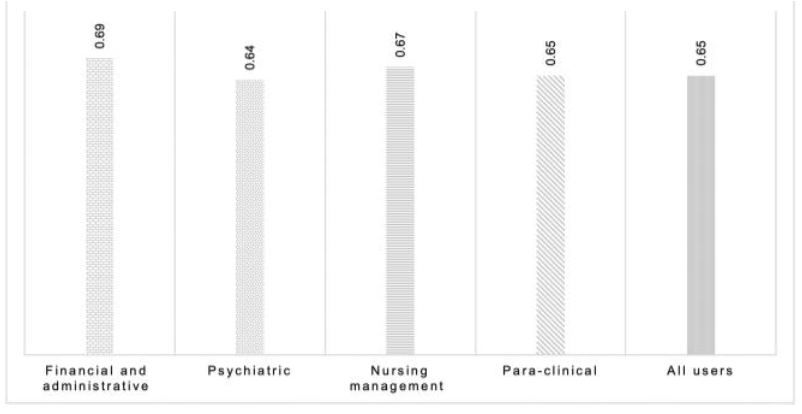
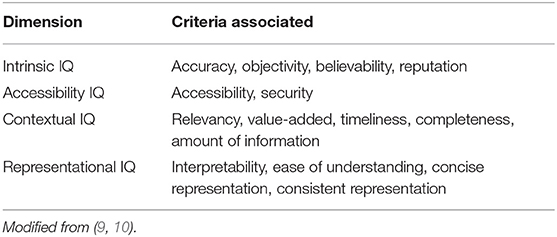
'''"[[Journal:Determining the hospital information system (HIS) success rate: Development of a new instrument and case study|Determining the hospital information system (HIS) success rate: Development of a new instrument and case study]]"''' | |||
A [[hospital information system]] (HIS) is a type of health information system which is widely used in clinical settings. Determining the success rate of a HIS is an ongoing area of research since its implications are of interest for researchers, physicians, and managers. In the present study, we develop a novel instrument to measure HIS success rate based on users’ viewpoints in a teaching [[hospital]]. The study was conducted in Ibn-e Sina and Dr. Hejazi Psychiatry Hospital and education center in Mashhad, Iran. The instrument for data collection was a self-administered structured questionnaire based on the information systems success model (ISSM), covering seven dimensions, which includes system quality, [[information]] quality, service quality, system use, usefulness, satisfaction, and net benefits. The verification of content validity was carried out by an expert panel. The internal consistency of dimensions was measured by Cronbach’s alpha. Pearson’s correlation coefficient was calculated to evaluate the significance of associations between dimensions. The HIS success rate on users’ viewpoints was determined. ('''[[Journal:Determining the hospital information system (HIS) success rate: Development of a new instrument and case study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 08–14:</h2> | |||
'''"[[Journal:Smart information systems in cybersecurity: An ethical analysis|Smart information systems in cybersecurity: An ethical analysis]]"''' | |||
This report provides an overview of the current implementation of smart information systems (SIS) in the field of [[cybersecurity]]. It also identifies the positive and negative aspects of using SIS in cybersecurity, including ethical issues which could arise while using SIS in this area. One company working in the industry of telecommunications (Company A) is analysed in this report. Further specific ethical issues that arise when using SIS technologies in Company A are critically evaluated. Finally, conclusions are drawn on the case study, and areas for improvement are suggested. Increasing numbers of items are becoming connected to the internet. Cisco—a global leader in information technology, networking, and [[cybersecurity]]—estimates that more than 8.7 billion devices were connected to the internet by the end of 2012, a number that will likely rise to over 40 billion in 2020. ('''[[Journal:Smart information systems in cybersecurity: An ethical analysis|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: July 01–07:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Mudge ScientificReports2018 8.png|240px]]</div> | |||
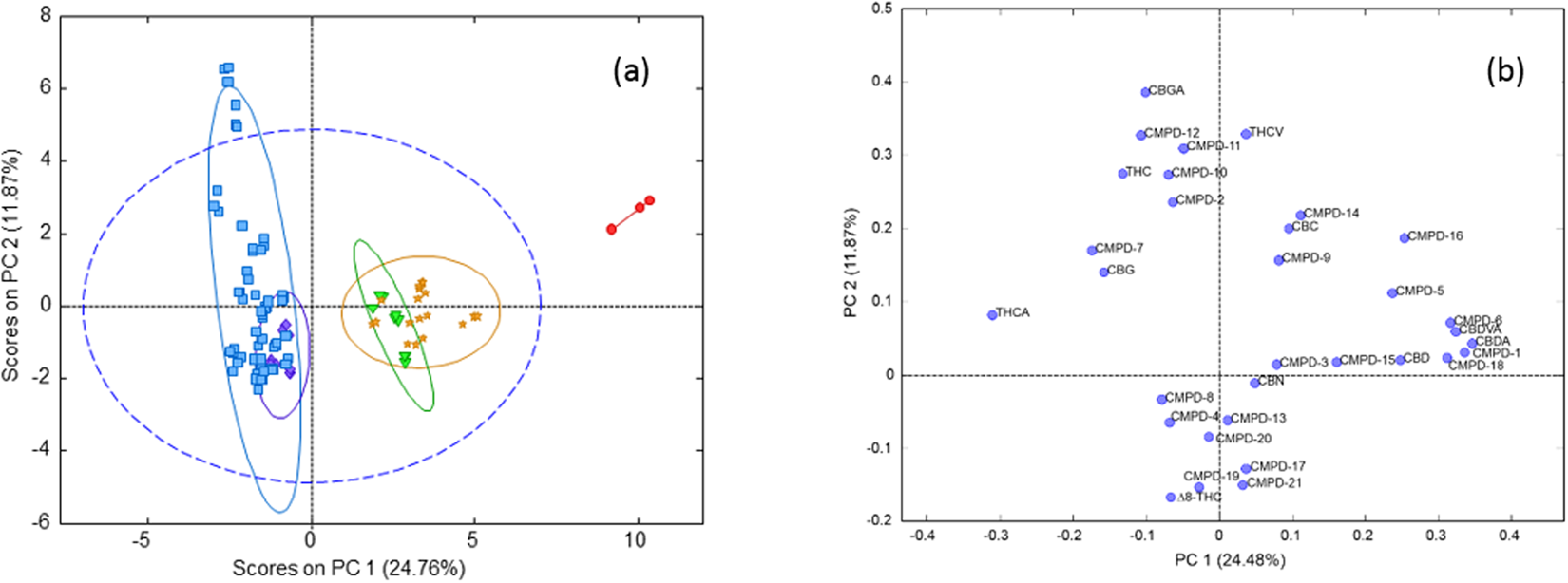
'''"[[Journal:Chemometric analysis of cannabinoids: Chemotaxonomy and domestication syndrome|Chemometric analysis of cannabinoids: Chemotaxonomy and domestication syndrome]]"''' | |||
''[[wikipedia:Cannabis|Cannabis]]'' is an interesting domesticated crop with a long history of cultivation and use. [[wikipedia:Cannabis strains|Strains]] have been selected through informal breeding programs with undisclosed parentage and criteria. The term “strain” refers to minor morphological differences and grower branding rather than distinct cultivated varieties. We hypothesized that strains sold by different licensed producers are chemotaxonomically indistinguishable and that the commercial practice of identifying strains by the ratio of total Δ9-[[wikipedia:Tetrahydrocannabinol|tetrahydrocannabinol]] (THC) and [[wikipedia:Cannabidiol|cannabidiol]] (CBD) is insufficient to account for the reported human health outcomes. We used targeted [[wikipedia:Metabolomics|metabolomics]] to analyze 11 known [[wikipedia:Cannabinoid|cannabinoid]]s and an untargeted metabolomics approach to identify 21 unknown cannabinoids. Five clusters of chemotaxonomically indistinguishable strains were identified from the 33 commercial products. Only three of the clusters produce cannabidiolic acid (CBDA) in significant quantities, while the other two clusters redirect metabolic resources toward the [[wikipedia:Tetrahydrocannabinolic acid|tetrahydrocannabinolic acid]] (THCA) production pathways. ('''[[Journal:Chemometric analysis of cannabinoids: Chemotaxonomy and domestication syndrome|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 24–30:</h2> | |||
'''"[[Journal:National and transnational security implications of asymmetric access to and use of biological data|National and transnational security implications of asymmetric access to and use of biological data]]"''' | |||
Biology and [[biotechnology]] have changed dramatically during the past 20 years, in part because of increases in computational capabilities and use of engineering principles to study biology. The advances in supercomputing, data storage capacity, and [[Cloud computing|cloud platforms]] enable scientists throughout the world to generate, analyze, share, and store vast amounts of data, some of which are biological and much of which may be used to understand the human condition, agricultural systems, evolution, and environmental ecosystems. These advances and applications have enabled: (1) the emergence of data science, which involves the development of new algorithms to analyze and [[Data visualization|visualize data]]; and (2) the use of engineering approaches to manipulate or create new biological organisms that have specific functions, such as production of industrial chemical precursors and development of environmental bio-based sensors. Several biological sciences fields harness the capabilities of computer, data, and engineering sciences, including synthetic biology, precision medicine, precision agriculture, and systems biology. These advances and applications are not limited to one country. This capability has economic and physical consequences but is vulnerable to unauthorized intervention. ('''[[Journal:National and transnational security implications of asymmetric access to and use of biological data|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 17–23:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab4 Wholey FrontPubHealth2019 6.jpg|240px]]</div> | |||
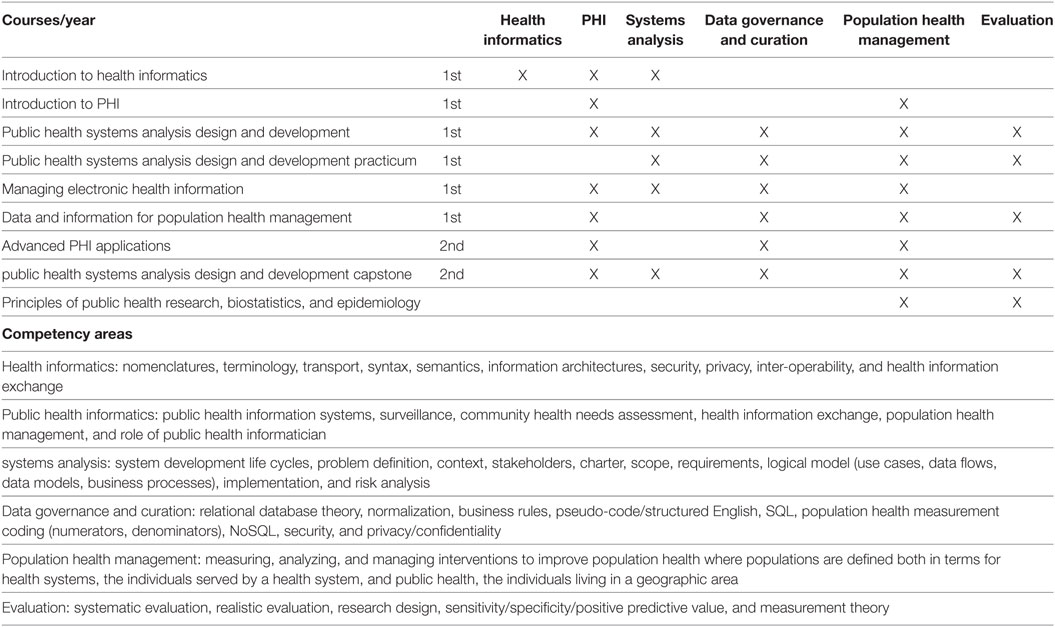
'''"[[Journal:Developing workforce capacity in public health informatics: Core competencies and curriculum design|Developing workforce capacity in public health informatics: Core competencies and curriculum design]]"''' | |||
We describe a master’s level [[public health informatics]] (PHI) curriculum to support workforce development. Public health decision-making requires intensive [[information management]] to organize responses to health threats and develop effective health education and promotion. PHI competencies prepare the public health workforce to design and implement these information systems. The objective for a master's and certificate in PHI is to prepare public health informaticians with the competencies to work collaboratively with colleagues in public health and other health professions to design and develop information systems that support population health improvement. The PHI competencies are drawn from computer, information, and organizational sciences. A curriculum is proposed to deliver the competencies, and the results of a pilot PHI program are presented. Since the public health workforce needs to use information technology effectively to improve population health, it is essential for public health academic institutions to develop and implement PHI workforce training programs. ('''[[Journal:Developing workforce capacity in public health informatics: Core competencies and curriculum design|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 10–16:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Schabacker FrontBioengBiotech2019 7.jpg|240px]]</div> | |||
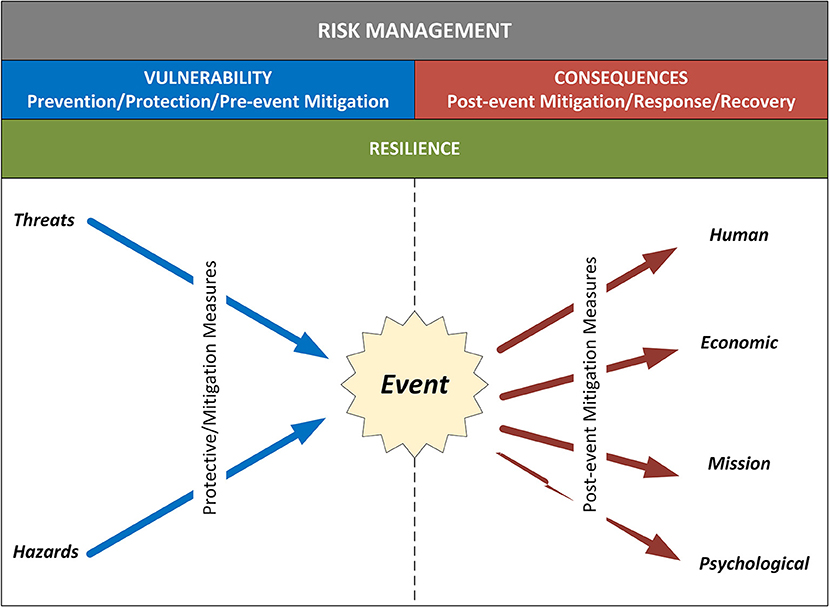
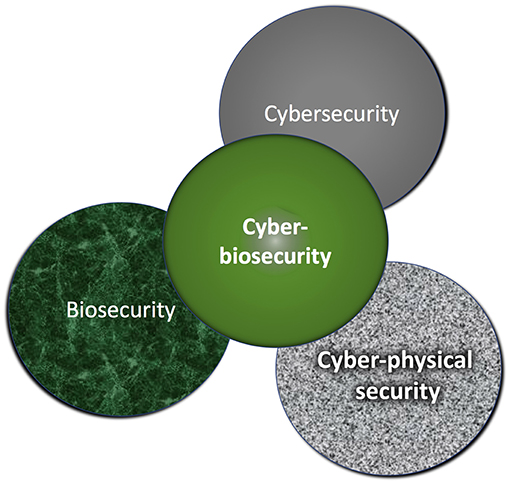
'''"[[Journal:Assessing cyberbiosecurity vulnerabilities and infrastructure resilience|Assessing cyberbiosecurity vulnerabilities and infrastructure resilience]]"''' | |||
The convergence of advances in [[biotechnology]] with [[laboratory automation]], access to data, and computational biology has democratized biotechnology and accelerated the development of new therapeutics. However, increased access to biotechnology in the digital age has also introduced additional security concerns and ultimately spawned the new discipline of cyberbiosecurity, which encompasses [[cybersecurity]], cyber-physical security, and biosecurity considerations. With the emergence of this new discipline comes the need for a logical, repeatable, and shared approach for evaluating facility and system vulnerabilities to cyberbiosecurity threats. In this paper, we outline the foundation of an assessment framework for cyberbiosecurity, accounting for both security and resilience factors in the physical and cyber domains. This is a unique problem set, yet despite the complexity of the cyberbiosecurity field in terms of operations and governance, previous experience developing and implementing physical and cyber assessments applicable to a wide spectrum of critical infrastructure sectors provides a validated point of departure for a cyberbiosecurity assessment framework. ('''[[Journal:Assessing cyberbiosecurity vulnerabilities and infrastructure resilience|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 3–9:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab2 Haugsbakken NordicJOfSciTechStud2018 6-1.png|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab2 Haugsbakken NordicJOfSciTechStud2018 6-1.png|240px]]</div> | ||
'''"[[Journal:What is the meaning of sharing: Informing, being informed or information overload?|What is the meaning of sharing: Informing, being informed or information overload?]]"''' | '''"[[Journal:What is the meaning of sharing: Informing, being informed or information overload?|What is the meaning of sharing: Informing, being informed or information overload?]]"''' | ||
Latest revision as of 15:36, 2 January 2024
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 - 2023 - 2024 |
Featured article of the week archive - 2019
Welcome to the LIMSwiki 2019 archive for the Featured Article of the Week.
Featured article of the week: December 23–31:"Building infrastructure for African human genomic data management" Human genomic data are large and complex, and require adequate infrastructure for secure storage and transfer. The National Institutes of Health (NIH) and The Wellcome Trust have funded multiple projects on genomic research, including the Human Heredity and Health in Africa (H3Africa) initiative, and data are required to be deposited into the public domain. The European Genome-phenome Archive (EGA) is a repository for sequence and genotype data where data access is controlled by access committees. Access is determined by a formal application procedure for the purpose of secure storage and distribution, which must be in line with the informed consent of the study participants. H3Africa researchers based in Africa and generating their own data can benefit tremendously from the data sharing capabilities of the internet by using the appropriate technologies. The H3Africa Data Archive is an effort between the H3Africa data generating projects, H3ABioNet, and the EGA to store and submit genomic data to public repositories. (Full article...)
|