LII:Next Generation Sequencing (NGS)/Pre-processing
Pre-processing
[edit | edit source]FASTQ files – discussion of the various quality encodings
[edit | edit source]FASTQ files extend sequence information in FASTA files by also including information about sequence quality. A FASTQ file typically consists of four lines.
- A line starting with @ and containing the sequence identifier
- The actual sequence
- A line starting with + followed by an optional sequence identifier
- A line with quality values encoded in ASCII space
As such the 2nd and 4th line must have the same length One such entry is given below showing one sequence "ATGTCT"...
@HWI-ST999:102:D1N6AACXX:1:1101:1235:1936 1:N:0: ATGTCTCCTGGACCCCTCTGTGCCCAAGCTCCTCATGCATCCTCCTCAGCAACTTGTCCTGTAGCTGAGGCTCACTGACTACCAGCTGCAG + 1:DAADDDF<B<AGF=FGIEHCCD9DG=1E9?D>CF@HHG??B<GEBGHCG;;CDB8==C@@>>GII@@5?A?@B>CEDCFCC:;?CCCAC
Here a quality value ranging from -5 to 41 is added to an offset and the resulting character is taken from an ASCII table. As such the whole data can be represented as text. Whilst Illumina made multiple changes to the quality format and eventually returned to almost Sanger encoding, the most important difference is whether the offset is 33 as in Sanger and Illumina v1.8 and later or 64 as in previous Illumina (and Solexa) formats. As you can see from the chart if you find any of the following characters: !"#$%&'()*+,-./0123456789: your offset must be 33 whereas any of the following characters KLMNOPQRSTUVWXYZ[\]^_`abcdefgh point towards an offset of 64. The above example is thus base offset by 33 as we find a 1 as first character. Also bear in mind that the @ and + signs are valid characters for quality so even if a line started by @ or + this could just be the beginning of the quality string.
See the quality chart below which is modified from the wikipedia article.
SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS..................................................... ..........................XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX...................... ...............................IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII...................... .................................JJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJ...................... LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLL.................................................... !"#$%&'()*+,-./0123456789:;<=>?@ABCDEFGHIJKLMNOPQRSTUVWXYZ[\]^_`abcdefghijklmnopqrstuvwxyz{|}~ | | | | | | 33 59 64 73 104 126 0........................26...31.......40 -5....0........9.............................40 0........9.............................40 3.....9.............................40 0........................26...31........41
S - Sanger and Illumina 1.8+, Offset 33, Quality range (0, 40) (!,I) X - Solexa, Offset 64, Quality range (-5, 40) (;,h) I - Illumina 1.3+ Offset 64, Quality range (0, 40) (@,h) J - Illumina 1.5+ Offset 64, Quality range (3, 40) (B,h) L - Illumina 1.8+ Offset 33, Quality range (0, 41) (!,J)
Presentation of the metrics used in QC
[edit | edit source]When you turn to quality control there are various metrics to consider.
Sequence Quality
[edit | edit source]The simplest is obiviously the quality score introduced in the FASTQ files above. As such it gives already a valid idea about base call quality. As often quality of reads degrades over the course of a sequence it is common practice to determine the average quality of the first, second, third,...nth base by just averaging over all reads in a file. Also to give some idea about the spread usually bar plots showing quantiles are given. This would give us an idea about what kind of trimming and quality filtering the data requires.
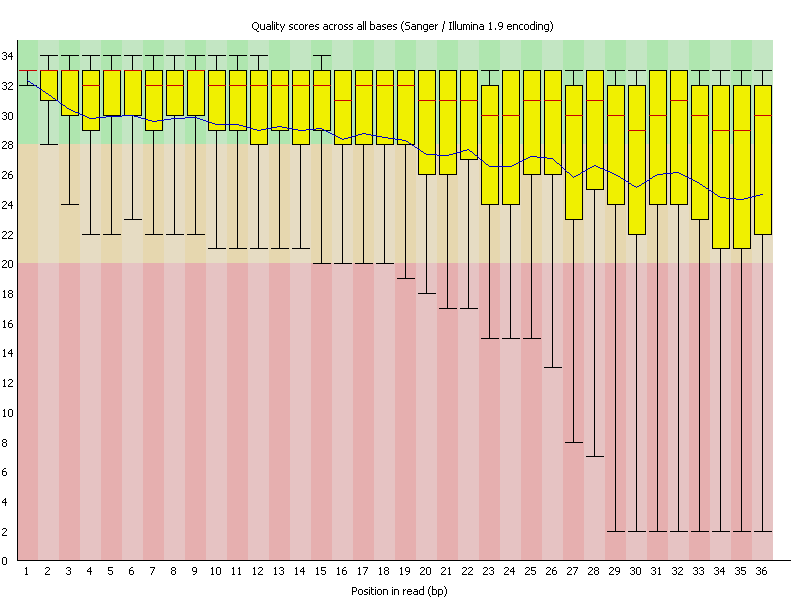
As an example here sequence data was investigated for quality using FastQC. As you can see the sequence reads are 36 bases long and the average sequence quality (depicted by the blue line) is steadily declining. In many new Illumina kits the sequence quality goes up a bit first before it steadily declines.
However instead of going over each base one can average the quality of each read instead and show a cumulative plot of the sequence quality of these.
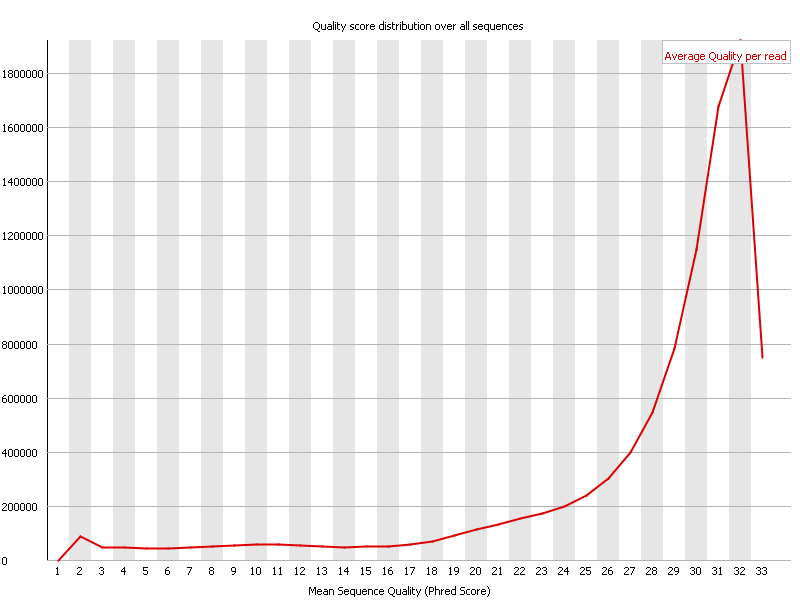
In the above screenshot one can observe that most reads have an average quality of 32. This is to be considered very good in general, however given that these reads are somewhat on the short side, it is probably at best an OK result.
Per Base Sequence Content
[edit | edit source]Another important metric is to look for base content at each position. Assuming the data is a random sample from the sequence space, at each position the contribution should be identical. Thus one needed to see straight lines. In reality it often happens that the first few bases might indeed show some erratic behavior, which could be due to non completely random primers. In the shown example however the reads are completely off. As you can see there is considerable bias in each base over the whole reads. In fact this bias is so strong, that you can read the overrepresented bases of the read.

As an example if you look at the last few bases you can read them as CTTGAAA-end of sequence.
Adapter sequence present or not?
[edit | edit source]If we now turn our attention to the overrepresented sequences in FastQC we can immediately figure out where this came from:
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCTCGTATGCCGTCTTCTGCTTGAAA | 1870684 | 19.446980406066654 | Illumina Single End Adapter 1 (100% over 33bp) |
| GAAGAGCTCGTATGCCGTCTTCTGCTTGAAAAAAAA | 95290 | 0.9906017065918623 | Illumina Single End Adapter 1 (100% over 28bp) |
Intro to errors and quality scores/encoding
[edit | edit source]As mentioned above the base caller assigns a quality score which is then available for each base. This gives the estimated reliability for this base. Please note that depending on your sequencing platform the typical mistakes are different. Illumina's most prevalent form of mistake is a nucleotide exchange whereas 454, Ion Torrent/Proton and other similar platforms have major issues with homopolymers such as AAAAAA where the correct number of As can often not be determined exactly.
Preprocessing Steps
[edit | edit source]Sequence Quality Trimming
[edit | edit source]In order to cope with lower quality data it is common to remove low quality bases. Typically one would remove lower quality bases from the e.g. the 3' end using a sliding windows approach as the per base quality gradually drops.
Alternative clipping strategies (Adaptor clipping)
[edit | edit source]In addition to removing lower base quality data, one would also remove adapters, PCR primers and other artifacts. In practice one would combine the adapter clipping with quality trimming approaches.
K-mer filtering/correction strategies
[edit | edit source]There are different ways to correct for base errors using kmer approaches because some errors can not be simply clipped off and even a very good quality value does not mean that a read is really error free. This is an important step prior to an assembly but potentially less crucial for alignments.
One basic idea is based on kmers in the read string. The original idea going back at least until 2001 (Pevzner 2001) generates a spectrum of kmers first, then kmers which are above a certain threshold (called solid) and kmers below this threshold are potentially arising from mistakes.
If a read is split into multiple kmers a single sequencing error will result in converting several overlapping kmers from strong to weak ones. An error correction step could now try to find the smallest number of changes required to make all kmers in the read strong.
Variants such as Quake also take the base quality into account to be better able to discriminate between low copy true kmers and high copy error kmers.
Digital Normalization and Partitioning
[edit | edit source]When considering especially RNA sequencing, it is well known that a normalization of RNA using molecular biology techniques (lab normalization) can help in providing better contigs and that a better general representation is achieved. This is because using lab normalization depletes common -or highly represented- sequences. Thus, when sampling from sequence space, after lab normalization it is less likely to find the previously very common sequences and thus more likely to find the previously underrepresented sequences. Apart from the advantage of having a higher likelihood to find underrepresented sequences there is the additional advantage that it is now less likely to find the same sequencing error multiple times in two or or more independent reads due to sheer oversampling. The latter makes it less likely that assembly software would erroneously create multiple contigs out of one true mRNA, due to these correlated SNPs. That said, lab normalization is neither easy and if it is outsourced it can be costly. Thus, one can instead use digital normalization. The basic idea is to downsample reads that have a lot of abundant kmwers. In addition this has the added benefit (to the ones above) that the number of reads to process becomes smaller, and thus it might be much more feasible (and faster) to assemble a transcriptome. One way to go about this is to use Titus Brown's tool set: http://ged.msu.edu/papers/2012-diginorm/
Paired end merging
[edit | edit source]A number of tools will take Illumina paired-end data and merge the reads if an overlap can be detected between them, potentially correcting errors by a taking the higher quality basecall at discrepant positions. This may improve assembler performance by reducing the data complexity, and may also improve the resulting contigs by removing erroneous data and improving the assembly of repeats. Tools to accomplish this include COPE and FLASH.
Removal of other undesirable sequences
[edit | edit source]Depending on the design of an experiment, there may be other sequences which are desirable to remove or mask from the reads prior to assembly. For example, if sequencing pools of BAC or cosmid DNA, it may be desired to remove most if not all of the vector backbone. Similarly, E.coli sequences will contaminate BAC or cosmid DNA preparations and could be removed in advance. Removing these post-assembly is an option as well. The PhiX control viral DNA is a common contaminant in Illumina sequencing data. Fast search tools such as SMALT can be used to map reads against a reference genome in order to identify those which should be removed.
Exercise
[edit | edit source]QC workflow (Data from machine -> pre-trimming quality plots -> trimming -> post-trimming quality plots)
[edit | edit source]A typical workflow might thus be to first get the data from the machine, and evaluate the typical quality plots as shown in the previous section. This gives a valid and important insight into the read quality and might potentially raise awareness about library preparation problems that might have occurred. After this problems have been identified and noted down, one would try to remove several errors by using trimming tools such as Trimmomatic to remove low quality bases from the sequence end and (potentially more importantly) to also remove remaining adapters etc from the reads. After having thus processed the reads, one would once again judge the quality to inform about remaining quality issues. As an example, even after removing known adapters from the sequences as in the above case, one might still see a per base sequence bias and would want to remove this bias or at least keep it in mind. We will discuss one exemplary workflow here.
The dataset
[edit | edit source]Download SRR074262 from the SRA (this is the example from above).
Fastq output analysis
[edit | edit source]Download FastQC (or analyze your data in RobiNA for similar plots). FastQC is relatively self explanatory. Open the FastQ file you just downloaded. FastQC will run through your dataset and you generate the plots shown in the introduction by clicking on the individual categories on the left hand side.
Adapter removal only
[edit | edit source]We will use Trimmomatic to simply remove adapters. java -jar trimmomatic-0.30.jar SE -phred33 SRR074262.fastq aclipped.fq.gz ILLUMINACLIP:TruSeq2-SE.fa:2:30:10 MINLEN:25 This tells trimmomatic that the quality encoding is phred 33 (modern Illumina) and it will store the results in the compressed file adapter_clipped.fq.gz. Finally it will use TruSeq3 adapters provided by trimmomatic.
Trimmomatic report
Input Reads: 9619406 Surviving: 7443332 (77.38%) Dropped: 2176074 (22.62%) TrimmomaticSE: Completed successfully
FastQC automatically recognizes gz files, so having the file compressed is ok. Indeed when we open the file aclipped.fq.gz in FASTQC the adapters have been removed.
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GGGGAGGAGAGAGCCATTGTTGAGGCGGCCATTGAG | 46910 | 0.630228505190954 | No Hit |
| AGGGGAGGAGAGAGCCATTGTTGAGGCGGCCATTGA | 21029 | 0.28252132244000405 | No Hit |
Adapter Removal and quality clipping
[edit | edit source]We will use Trimmomatic to remove adapters and use a sliding window approach to remove lower quality bases at the end
java -jar trimmomatic-0.30.jar SE -phred33 SRR074262.fastq aclippedtrim.fq.gz ILLUMINACLIP:TruSeq2-SE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:25
Input Reads: 9619406 Surviving: 6513310 (67.71%) Dropped: 3106096 (32.29%) TrimmomaticSE: Completed successfully
Read correction
[edit | edit source]Links to useful tools for preprocessing
[edit | edit source]Links to resources used in this chapter
[edit | edit source]Links to other QC tools e.g. FASTX toolkit and spectral alignment for error correction
[edit | edit source]
Notes
This article is a direct transclusion of the Wikibooks article and therefore may not meet the same editing standards as LIMSwiki.









