Journal:Using OpenBIS as a virtual research environment: An ELN-LIMS open-source database tool as a framework within the CRC 1411 Design of Particulate Products
| Full article title | Using OpenBIS as a virtual research environment: An ELN-LIMS open-source database tool as a framework within the CRC 1411 Design of Particulate Products |
|---|---|
| Journal | Data Science Journal |
| Author(s) | Plass, Fabian; Englisch, Silvan; Zubiri, Benjamin A.; Pflug, Lukas; Spiecker, Erdmann; Stingl, Michael |
| Author affiliation(s) | Friedrich-Alexander-Universität Erlangen-Nürnberg |
| Primary contact | Email: michael dot stingl ay fau dot de |
| Year published | 2023 |
| Volume and issue | 22 |
| Article # | 44 |
| DOI | 10.5334/dsj-2023-044 |
| ISSN | 1683-1470 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://datascience.codata.org/articles/10.5334/dsj-2023-044 |
| Download | https://datascience.codata.org/articles/1500/files/655de35843b0d.pdf (PDF) |
Abstract
The transformation of existing technologies and consequent use of new digital technologies not only have a substantial impact on society and companies, but also on science. Analog documentation and research, as we have known it for centuries, will eventually be replaced by intelligent, more FAIR (findable, accessible, interoperable, and reusable) digital methods ands systems. In addition to the actual research data and results, metadata now plays an important role not only for individual, independently existing projects, but also for future scientific use and interdisciplinary research groups and disciplines. The solution presented here, consisting of an electronic laboratory notebook (ELN) and laboratory information management system (LIMS) based on the openBIS (open Biology Information System) environment, offers interesting features and advantages, especially for interdisciplinary work. The Collaborative Research Centre (CRC) 1411 "Design of Particulate Products" of the German Research Foundation is characterized by the cooperation of different working groups of synthesis, characterization, and simulation, and therefore serves as a model environment to present this implementation of openBIS. OpenBIS, as an open-source ELN-LIMS solution following FAIR principles, provides a common set of general entries, with the possibility of sharing and linking (meta-)data to improve the scientific exchange between all users.
Keywords: open science, research data management, databases, ELN-LIMS, interdisciplinary work
Introduction
Digital transformation is a key challenge that impacts our entire society. The main players here are companies that use intelligent information technologies (IT) and networks for machines and processes. Starting with flexible production via modular, changeable production processes and moving to customer-specific solutions and products requires the help of sophisticated data acquisition, processing, and analysis.[1][2] Further examples of digital transformation include automation processes, machine-to-machine communication, internet of things (IoT) process implementation, or even augmented-reality-based workflows.[1][2][3][4]
However, not only companies are subject to this change, but also government institutions (eGovernment)[5] and science itself.[6] Thus, the topic of "open science" has become a growing movement.[7] Open science has been supported in the European context of the Open Research Data and Data Management Plans of the European Research Council (ERC), established by the European Commission for almost five years. However, the ERC has been promoting the causes of open science since 2007.[8] This also has manifested in other ways, as, for example, open access publications from funded projects that have already become mandatory to a certain extent. Exemplarily, the DFG (German Research Foundation) supports infrastructure projects within Collaborative Research Centres (CRCs), which are long-term university-based research institutions established for up to 12 years, and whose funding objective is to establish powerful information systems for research in a holistic perspective.[9] Accordingly, new infrastructure on a national (i.e., Germany's National Research Data Infrastructure; Nationale Forschungsdateninfrastruktur or NFDI) and European level (i.e., the European Open Science Cloud or EOSC)[10][11] have been established, fostering the subject of research data management, data publications, and open science. Nevertheless, the topic of open science includes more than just the public provision of data in the context of open-access publications; it also includes the approaches of open methodologies, sources, and data. The necessary implementation and representation of good scientific data management, data quality, and stewardship (data governance) are tremendously important.[12][13][14][15] The resulting benefits can be measured directly, such as in terms of improvements in process efficiency or cost and risk reductions, and indirectly, such as increased acceptance, perception, and trust.[12][13][16]
This paper presents an implementation of openBIS, an electronic laboratory notebook (ELN) and laboratory information management system (LIMS) to support open science broadly, including data management, handling, storage, and publishing within a scientific laboratory environment.
Current state of research data management
FAIR as part of research data management
One of the cornerstones of this overall research data management (RDM) is the FAIR principles, which encourage research objects to be more findable, accessible, interoperable, and reusable. The emphasis placed on and the growing awareness of FAIRness is, however, more than just an essential duty that public funding agencies impose on research. Moreover, it is the key to conduct knowledge discovery, innovation, and information transfer, as well as the subsequent integration and reuse of research objects by the scientific community.[15] Events such as the global COVID-19 pandemic demonstrate the need for, and the overall benefits of, making data available online in a reusable fashion.[17][18] This leads not only to efficient research and increased innovation, but also to fair and transparent use of public funds and tax capital, as well as increased visibility and scientific reputation and reliability, to name just a few benefits of open science.[19]
The FAIR principles propose that all scholarly output should embody the characteristics of being findable, accessible, interoperable, and reusable. While these principles provide guidance on the expected behaviors of data resources, their practical implementation has been subject to varying interpretations. As the support for these principles has grown, so has the diversity of interpretations surrounding their application.[11] FAIR principles recognize the need for data accessibility under defined conditions, but do not necessitate complete openness. While transparency and clarity are required for accessing and reusing data, restrictions can still remain based on privacy, security, and competitive concerns. FAIR promotes a balanced approach that allows diverse participation and partnerships while ensuring the availability of data within specified guidelines.[11]
Data repositories and data publications
Data repositories are a key component of the digital transformation of science. Well-known examples are the commercial data repository service Figshare, open-access archives like arXiv, or platforms like Dataverse[20], EUData[21], and Zenodo, which is maintained by CERN and funded by the E.U. Commission. In fact, most of the known repositories already consider the high-level FAIR principles. In the case of Zenodo, the uploaded data is provided with a digital object identifier (DOI) and can optionally be published as open-accessible and viewable. This, in turn, leads to simplified findability, accessibility, and usability for the scientific community, as well as to the quotability of individual datasets, whose content no longer has to lead directly to a complete publication. This means that even data, methods, or code that initially received little attention can now be found by the general public and are not lost.
Despite all of this, justified doubts exist regarding the open science policy. Especially, there are questions and concerns about the security of data against external interference and possible compliance and regulatory requirements, especially in healthcare, such as the protection of relevant patient data. These issues circling around the subject of data sovereignty need to be clarified during the planning of and before introducing digital technologies such as data management or cloud-based systems.[22][23]
Good research data management does not start with the publication and archiving process of the work or the (meta-)data, but with their initial collection. This is because not only content-related data/information is of importance in the sense of RDM; metadata is as well. Metadata describes data or in general (additional) information about the described data(set). Metadata-specific information can be the authors, the creation time/date, or the type of the dataset, as well as the DOI of the dataset. In fact, many distinct types of metadata exist, including descriptive, structural, administrative, and process data.
ELNs and LIMS
The scientific questions, choices of experimental procedures, materials and methods, data analyses, and interpretation of research and its results were traditionally recorded in detail in paper-based laboratory notebooks.[24] Not only is this approach incomprehensible today, since most data are generated electronically or stored as code on a network anyway, but this concept vehemently contradicts the overarching principles of FAIR, as well as open science in general. Neither are the data easy to find, nor are they accessible or usable by scientists outside the local physical system in which they are stored. Inevitably, the use of paper-based notebooks should be avoided, and a shift made to ELNs and LIMS.
The combination of an ELN with a LIMS allows research labs to facilitate the documentation and management of laboratory processes and data. An ELN is used for capturing and organizing experimental data, while a LIMS supports the management of laboratory resources, sample tracking, quality assurance, and other laboratory functions. Together, ELN and LIMS provide a comprehensive platform for efficient and secure management of laboratory information, promoting compliance with best practices and regulatory requirements.[24][25][26] ELNs can play a major role in a successful RDM effort. In this way, a continuous workflow under FAIR conditions can be guaranteed from the very beginning, starting with the collection of data, the use of the data by oneself or the research group and other researchers, the publication of the data, and, finally, a superior archiving, for example of data repositories like Zenodo or RADAR.[27] Further advantages of an ELN system include[24]:
- the easy and metadata-based collection of information, improving their shareability;
- an ensured long-lasting data storage on a secure server;
- simplified accessibility via a global or local network;
- greater archiving possibilities on open data repositories; and
- self-implementable applications with the system.
Currently available project management tools connecting classical, collaborative project and data management efforts include platforms like OSF.[28] Classical ELN systems range from commercial applications like CERF[29], Benchling[30], and labfolder[31] to open-source options like Chemotion ELN[32], eLabFTW[33], and openBIS.[34]
Current situation, methodology, and strategy
Current situation from an interdisciplinary point of view
The scope of open science and RDM, as well as a clear and well-defined data stewardship and governance, has been clearly recognized from the perspective of the DFG-funded Collaborative Research Centre (CRC) 1411 Design of Particulate Products. However, an ELN-LIMS is currently lacking for the use within an interdisciplinary field involving engineering, materials science, natural sciences, mathematics, theoretical modeling, and simulation. Furthermore, commercial systems are generally not recommended due to their possible financial conditions, in addition to a thereby complementary open science policy. Open-source products like Chemotion ELN are, again, too subject-specific (in this particular case, chemistry), which would lead to a less beneficial impact on interdisciplinary work and research, such as lower usability, and, thus, in the end, also acceptance in other subject groups. On the other hand, the openBIS system of the Department of Biosystems Science and Engineering and Biology of the ETH Zurich[24][35] shows promising features that are worth a closer look, even though this system was designed primarily for biological and medical disciplines and is not only used by academic institutions, but also by companies in the industry sector.[35]
Accordingly, we describe the implementation of our openBIS system in several subsections, starting with the basic principles and working features of openBIS, continuing with the goals and requirements of researchers within the CRC and the scientific community for a well-functioning ELN tool, and ending with its implementation and usage. openBIS is, besides other ELN-LIMS examples, a good starting point and framework to foster cooperation via a digital environment using an ELN and by fulfilling the requirements by the DFG and NFDI consortia, such as FAIRmat (for materials science, physics, chemistry, and mathematics) with respect to data management, handling, storage, and publishing. In addition, we also want to share our experiences in further developing the system and its implementation and daily use.
General overview and structure of openBIS
Before we shed light on how openBIS can help us with a successful implementation of a beneficial RDM, we first clarify what openBIS is, how it works, and what technical requirements need to be met. OpenBIS is an open-source platform that functions both as an ELN and LIMS. Developed by ETH Zurich, openBIS provides an open-source database for research laboratories, especially designed and implemented in and for life sciences.[24] The goal was to build a simple and efficient, yet comprehensive ELN-LIMS system that meets the daily needs of a research institution. Everyday things like storage of materials; instrumental setups and devices; acquisition, description, and processing of large amounts of data; and sharing of research with users and scientists within the openBIS system should be possible.[35]
However, questions arise regarding what kind of technical conditions are needed to build openBIS within a research group, whether openBIS is scalable, and whether such a system in the area of a scientific network (E.U. or DFG fund)—or even a comprehensive database within an entire university—is conceivable at all. All of these questions are important to know before starting with an ELN-LIMS and will be discussed.
The history of openBIS and its platform, on which it is based, started in 2007 and is still actively maintained, nowadays by the Scientific IT Service Team of the ETH Zurich. openBIS requires a modern Unix-like operating system (OS), for instance Linux systems. However, openBIS can be run on virtual machines and docker containers and therefore is mainly platform-independent. A very interesting and detailed look on the general technical background of openBIS is provided by the developers.[35] However, shortly summarized, openBIS has one or several data store server(s) (DSS) and an application server (AS). On the AS, data provenance actions like metadata handling are conducted, while on the DSS the raw data is managed. While on the front end user access is facilitated via a web browser, the AS uses a relational database management system (RDBMS) to generate persistent information like index information about all data sets, and the data itself is covered and stored within the (several) DSS system(s). The latter is responsible for creating, querying, and visualizing data while they are mediated by the AS.[35] At the application side, the ELN-LIMS system is accessible via browser-based tools (the recommended ones are Chrome, Firefox, and Safari) and is reachable from any electronic device and operating system.
Moreover, as part of our INF project of the CRC 1411 (see the funding information in the acknowledgements), we want to further extend the concept of ELN-LIMS to a more virtual research environment in the future (see the next section for further information). Our goal is for VREs to be collaborative and present requirements-tailored tools that support web-based research environments.[36][37][38] The DFG defines “Virtuelle Forschungsumgebung” (a literal translation of "Virtual Research Environment") as a platform for internet-based collaborative working that enables new ways of collaboration and a new way of dealing with research data and information.[39]
Results
Features of openBIS as an ELN-LIMS system within CRC 1411
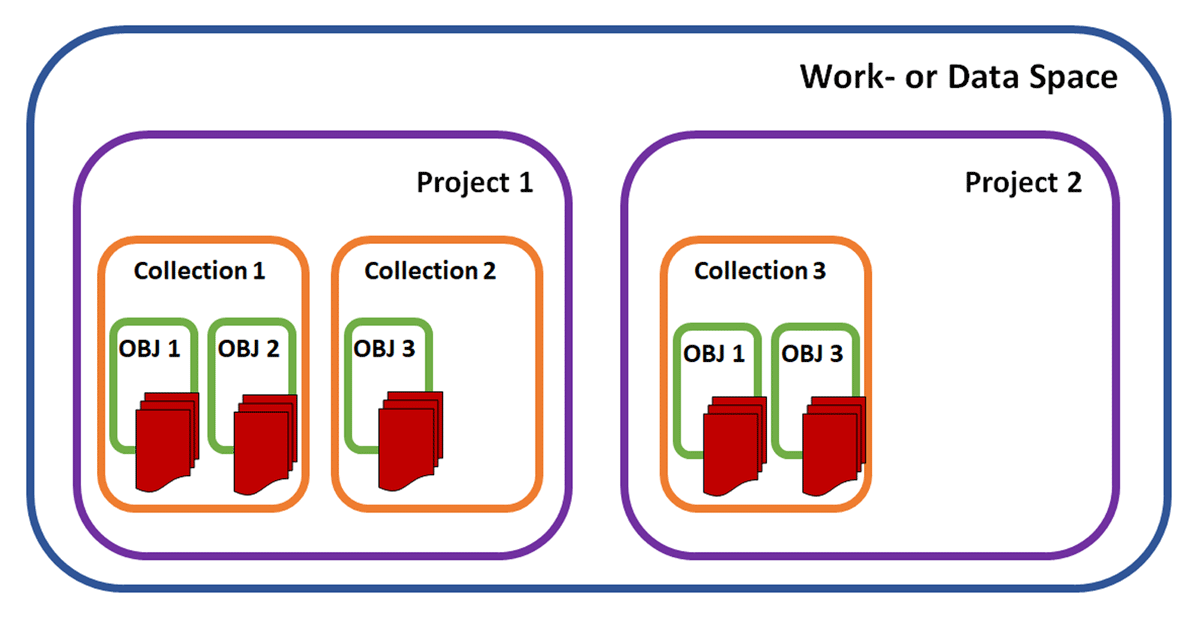
As openBIS is accessible for users via a web app, and even from outside the university network, access to the user’s own data and data provided by others is guaranteed always and everywhere. It is not only crucial to be able to create and implement (meta-)data, but doing so further enables the function that every user can permit access to its projects to selected users. This covers different roles (e.g., observer, user, admin) with different rights like reading, writing, or even deleting. Here, the authorization process lies completely in the hands of each user and can be adjusted independently, with respect to other users within the network (i.e., role-based) or other projects by the same user. In general, openBIS has a predefined hierarchical structure, which is divided into several levels. The first and main level is the "Work" or "Data Space" level, which, in addition to general rights and access, primarily contains all projects. The second level, "Projects," pertains to the projects themselves. These, in turn, have "Collections" (third level), which consist of different "Object Types" (fourth level), with or without datasets (see Figure 1) of type "Experimental Step," "Entry," or "Instrument."[35] This is also valid for the Collections level. In fact, the first and second level contain a persistent identifier with one specific path using the nomenclature “/user(workspace)/projectx.” Collections and Objects extend the identifier by a unique code. However, these are moveable between Projects and Spaces. In the example of Figure 1, we can move Collection 1 with all its entries to Project 2, which can be shared, but the Projects and Spaces are not moveable.
|
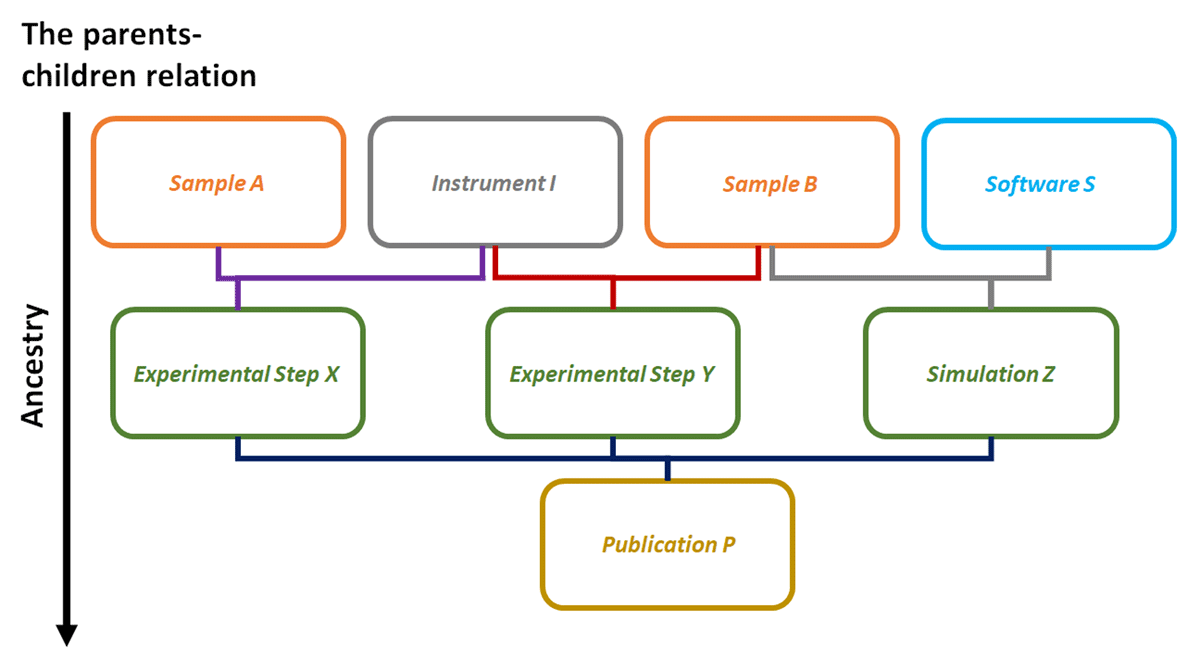
There are no limitations on the maximum number of users working on one openBIS ELN system. The only limitations are connected to the available and used server resources. It can also link data on samples, materials, instruments, and experiments. Here, openBIS features the parent-children relationship model (see Figure 2 for further detail). This means that every created Object type is logically interconnected between other Object types. As an example of our CRC, synthesis and characterization groups are trying to develop specific and well-defined particles considering different synthetic samples, materials, and processes. To ensure that the creation, modification, or deletion of any electronic records is traceable, computer-generated and time-stamped audit trails are used to record the date and time of any user interventions. As an example, to create an Experimental Step X and Y (see Figure 2), the researcher/user requires a different amount of Samples A and B, and one specific Instrument I. Moreover, for a specific Simulation Z, one uses Sample B and Software S. Here, Sample, Instrument, Software, and the Experimental Step and Software, or later on Publication, are all different created Object types that are accessible and existing for every user of our openBIS system. Furthermore, all Object Types within our system are classified into three different categories and differ if they are generalized entries (Entry and General Type), experimental procedures or analysis (like Experimental Step or Simulation), or properties and (real existing) objects (like Instrument or Software). In our basic examples, it is valid that Sample A and B, Instrument I, and Software S are all parents of the Experimental step X, Y, and Simulation Z, respectively, which are automatically the child(ren) of the prior Object types. If we now create an Object type Publication P, which is linked and fed via our Experimental step X, Y, and Simulation Z, then, accordingly, P is the child of X, Y, and Z, and vice versa. This results in a clear and well-understandable line of ancestry, which fulfills the FAIR concept. In our example, even after publishing (Publication P), one can clearly find and reuse (meta-)data of the corresponding project. Moreover, one does not have to create object types (and their underlying [meta-]data) multiple times, as one can reuse them if needed. This reduces redundancy of (meta-)data that is used across Spaces, Projects, and Collections, and saves storage and data maintenance time. Additionally, in an interdisciplinary environment, multiple groups utilize a common set of Object types, which would otherwise appear in a redundant manner in different fields. This can save a lot of time and lead to the possibility to create and implement a more comprehensible laboratory and research system.
|
OpenBIS can also be used for administrative and lab-specific workflow processes. To understand how to do this, we have to take one step back and consider how openBIS works from each user’s point of view. First, every user possesses its own Workspace, which can be compared to its own desktop or computer storage. Within this Workspace, projects, metadata, and other objects can be created and adjusted without necessarily being connected to other users. This is possible using the Manage Access option, a function that allows users to simply grant access to those users within the system who should be allowed to read, write, and/or delete information in your project. All permitted users will appear as folder icons below the own Workspace folder icon. However, one researcher or user is connected to a specific working group or institute containing several other researchers/users. Finding a way to bring the entire working group together without repeatedly pressing the Manage Access button and effectively defining user roles is a key consideration. Especially for general administrative or instruments within the institute, which are accessible for every researcher of the corresponding group, creating for example several Instrument object types is not only time-consuming, but it also holds the danger of creating different metadata for the same, for example, instrument. This would result in problems with the FAIR principles.
Moreover, openBIS includes an additional working area called Inventory, which serves as a "common work environment" for user-groups to access information and data stored within this section. This area houses data and information that is universally relevant or of interest to all users or larger groups, and it contains spaces for the CRC, such as an openBIS on-boarding or a common folder. Furthermore, we have made further adjustments to the Inventory section by implementing sub-Inventories that are specific to the working groups/institutes within our CRC.
These different spaces, including the Inventory and our institute-based Inventories, provide fields for collaborative projects and administrative workflows, offering a general and overall usage area, along with a sub-Inventory containing commonly used, institute-dependent instruments and materials, among others. With a collaboratively-used Inventory, each user has access to collections of items like Instrumentation, where necessary object types (e.g., Instrument I1, I2, …) are created once and can be utilized by any user within the workgroup (those with access permission to the specific folder of the Instrumentation section). The same concept applies to collections of samples or documents/protocols that are employed in large collaborative projects.
By following the principle of the parent-child relation depicted in Figure 2, we ensure that instruments (e.g., Instrument I from our example) within an Instrumentation folder can now be used for tracking as part of the Inventory folder, while avoiding redundant information and directly providing the required documentation. This makes it possible to combine a classical working group environment with interdisciplinarity and FAIR data management within an overall framework.
OpenBIS as a multi-institutional framework within a material science-based environment: Adjustments and experiences
This multi-institutional framework can now be implemented and customized to specific user needs. Via an additional overlay (Core UI), the implementation of different object types (from simple general types to specifically customized types) is possible. The implementation process will involve questions regarding the number and types to be implemented. Whether more specific and fine-tuned types are preferred or not needs to be clarified in advance. In our case, we shifted from specific object types (as the pre-defined openBIS system for a bio-medical environment provides with, e.g., bacteria, yeast, etc.) to more generalized object types such as sample or simulation, which are usable in an interdisciplinary environment by multiple groups at once. We decided to use openBIS as foundation or "hollow" version, while winnowing object types that exist by default but may be not required in our digital illustration of our CRC-based workflows, and to develop new ones using our own CRC-based ontology, accordingly. To achieve this, we decided to form a pilot group for all users within the CRC 1411 about six months before handing the openBIS system over to the entire group. Since our interdisciplinary working environment consists of different disciplines—from pure particle synthesis to their characterization, analysis, simulation, and theoretical optimization—we invited about 15 people from the different working groups/disciplines and implemented, discussed, and modified our preliminary system up to this point, so that everyone was satisfied with the result. It turned out that implementing dozens of different and fine-tuned object types for each user, while appearing functional at first sight, also brings some disadvantages, because while each object type is visible in the drop-down menu, not every object type is used by every user. For example, theoretical scientists, who use mathematical models to simulate and predict physical properties of potential nanoparticles, rarely use synthesis-based object types like General Protocol or Experimental Step. This means that any user from a particular discipline will only stick to the types that are useful for their own research. Furthermore, since each type is always present in the system’s drop-down menu, it quickly becomes cluttered and confusing, leading to poorer usability and, consequently, a less accepted package. Therefore, we finally decided to use only eleven object types, two of which are already preset by the developers (Entry and Experimental Step), eight of which contain specific (meta-)data, and one which is defined as an overall or general object type (General Type).
Meeting requirements (such as scientific or administrative needs) that may exist within the same discipline can be a challenge. Again, our implemented multi-institutional framework could help. Now, since each workgroup/environment has its own "space" within the overall openBIS system, it is possible to implement specific "workgroup-defined object types." For example, in our CRC 1411, synthesis and processes can vary between workgroups, so implementing finely tuned objects, which can only be used by users within that workgroup or users with access privileges, is preferable. Furthermore, there are no negative effects, such as usability on interdisciplinary work or traceability of (meta-)data, since, within the entire openBIS system (and users with access rights), the new (and now workgroup-specific) types can still be viewed. As a result, this leads to a balanced combination of a universal and well-usable openBIS data management system and a well-tailored work environment.
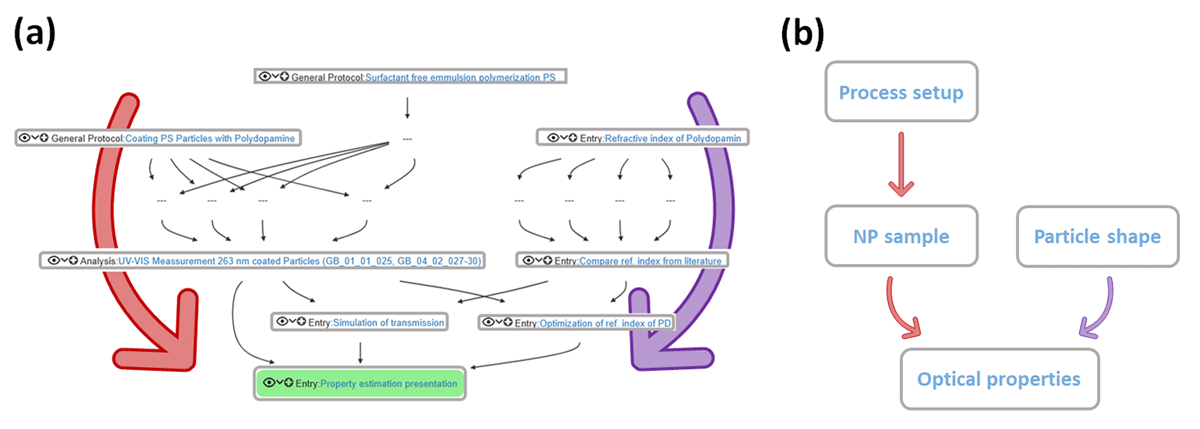
Now each user can edit and share their own data, both scientific and non-scientific in nature/origin, with themselves or with other users within the openBIS system by creating and storing data and metadata that can be exported in turn. An exemplary workflow, in which data are shared and exchanged among CRC 1411 users, is illustrated in Figure 3. The reasons for this can be quite different: more and more journals are demanding that special attention must be paid to open science and research data management, for instance, by using a data repository to which publication-specific (meta)-data must be uploaded and made freely available to all. In some journals, for example, it is common to prepare a data availability statement when submitting the publication. This statement includes information about the data itself, where to find it, an identifier for the data (i.e., DOI or persistent identifier), and how to access the data. The function used for re-exporting stored and listed (meta)data in openBIS is the Export Metadata & Data function. The openBIS server now plays the role of a kind of e-mail distributor. That is, openBIS exports the project folder selected by the user as a .zip file, which now contains all (meta)-data, as .txt, .doc, .html, and .json files, as well as the introduced structure by the researcher and the system. In fact, the system uses a terminology adapted for our workflows within the CRC. This means that the default syntax and terminology of our openBIS system, along with the modifications and additions made to display workflows for various research areas within our CRC, will be included in the .zip file. The management of the researcher’s data will therefore be displayed in the folder as well. Since each user has its own account in openBIS, the user can send the exported file to others directly to their e-mail inbox via a download link, with no data size limitations, by entering their e-mail address. This .zip file can now be attached to a publication as Supplementary or Supporting Information, or uploaded to a data repository, and the DOI obtained can be noted in the actual publication. This closes the entire research data management data lifecycle, from project inception through data production and analysis to long-term archiving. Moreover, as stated above, the pre-structured organization of your project(s) on openBIS (and via the Object types) stays the same after export and may be re-imported by writing a parser to another ELN system, such as openBIS. That means that no additional structuring process is needed. Furthermore, after implementation of the gathered publication or research papers that have been published under the CRC 1411 roof, we track all of them via openBIS now as well.
|
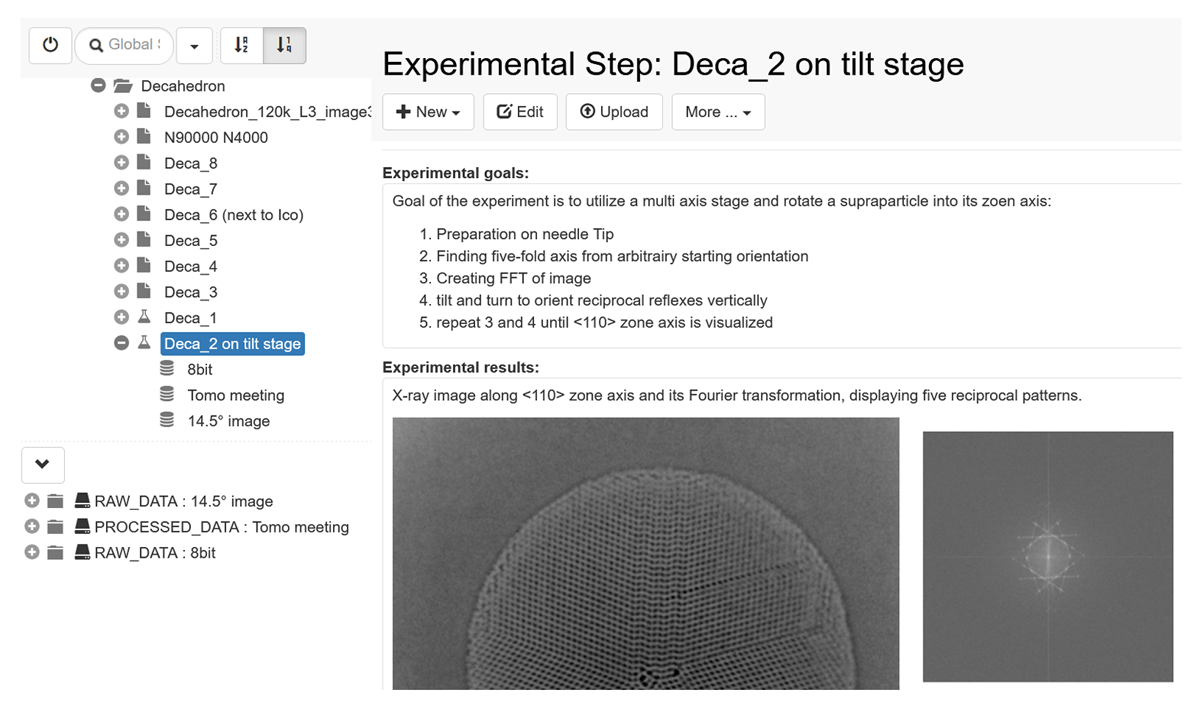
As mentioned at the beginning, the ELN-LIMS system openBIS@CRC is designed to adhere to FAIR principles. This is evident in various aspects of the system. Firstly, the data stored in the openBIS database can be easily found through persistent identifiers and a search function. Additionally, the data is easily accessible via the internet without the need for a virtual private network (VPN), allowing users to access the webapp from anywhere; the webapp graphical user interface (GUI) is illustrated in Figure 4. The system is also interoperable, allowing external and internal scripting to interact with the data. Moreover, the data and metadata can be exported, making it reusable. Alongside the FAIR principles, the system also promotes good scientific practice and follows the data management lifecycle, covering aspects such as documentation, tracking of projects and data, and archiving. Collaboration is facilitated through role management and the availability of separate workspaces for individuals and working groups within openBIS.
|
Our current openBIS system has been up and running since end of December 2021, but we are still testing new features and making minor and major changes. Following the rollout of the "final" version of the openBIS system to our users within the CRC 1411, we have organized several introductory events within our iRTG (integrated Research Training Group). In addition, we host monthly support meetings where questions or comments can be brought forward, and support is offered. Furthermore, we did not want to develop an exclusive data management system, so young research students, for instance in the context of their Bachelor’s or Master’s thesis, will have access as new users to get more hands-on work in with research data management. In addition, the purchase of lab notebooks and tablets has increased the adoption of openBIS, as data creation, processing, and sharing within the labs is now very fast and easy. The use of paper-based laboratory notebooks has been decreasing significantly over time.
Our CRC aims to expand the capabilities of the ELN-LIMS concept in the future. As described earlier, our vision of a virtual research environment includes integration of data repositories like Zenodo, enhanced visualization options including augmented/virtual reality technologies, and post-processing tools for stored raw data. This can be achieved through Python scripting using openBIS’s internal application programming interface (API), enabling the implementation of scripts and other post-processing functionalities. The flexibility and range of possibilities offered by openBIS played a significant role in our decision to select this ELN-LIMS for our CRC.
One specific example of collaboration and post-processing within our CRC involves the implementation of a script for color calculations of nanoparticles. This script will assist our synthesis groups in developing nanoparticle synthesis routes even before they begin their experiments. By utilizing the openBIS server, the synthesis groups can directly access the theoretical calculations typically performed by our theory groups, eliminating any time delays. This iterative process of development, deployment, implementation, and utilization accelerates research within our CRC and highlights the value of the openBIS ELN system. Furthermore, we are planning to integrate a Jupyter Hub system with our openBIS@CRC system in the future, providing researchers with additional post-processing and scripting options within our environment.
Conclusions
We demonstrate an ELN-LIMS solution that addresses the challenges and needs of multiple disciplines within a technical and natural science faculty on the openBIS system. In fact, the versatility and modifiability of the system formed the basis for a system for multiple working groups of the CRC 1411 Design of Particulate Products that meets the documentation needs of groups focusing on synthesis, characterization, and simulation, and, in particular, supports collaboration between different disciplines within the CRC. The structure was developed and adapted via several steps and provides both a common set of Object types and the ability to include more specific ones for individual working groups without disrupting common workflows. As a result, we discovered that the balance between providing users with flexibility and customization in an ELN-LIMS system and maintaining a commonly used and understandable interface, particularly concerning the number of object types, is delicate and requires careful consideration. Indeed, both inadequate and overwhelming design and functionality lead to a reduction in potential use by researchers and, therefore, require accurate design and communication between users and the administrators of the ELN-LIMS, especially in a highly interdisciplinary project like the one at our CRC. Ultimately, this ELN-LIMS could serve as a template solution for other similarly structured collaborative research centers or research groups.
Abbreviations, acronyms, and initialisms
- API: application programming interface
- AS: application server
- CRC: Collaborative Research Centre
- DFG: German Research Foundation
- DOI: digital object identifier
- DSS: data store server
- ELN: electronic laboratory notebook
- EOSC: European Open Science Cloud
- ERC: European Research Council
- FAIR: findable, accessible, interoperable, and reusable
- GUI: graphical user interface
- IT: information technology
- IoT: internet of things
- iRTG integrated Research Training Group
- LIMS: laboratory information management system
- NFDI: National Research Data Infrastructure (Nationale Forschungsdateninfrastruktur)
- OS: operating system
- RDBMS: relational database management system
- RDM: research data management
- VPN: virtual private network
Supplementary materials
- dsj-22-1500-s1.zip (2.37 MB): The aim of this research article is to show not only what openBIS can do in the interdisciplinary research environment, but also how metadata and data are ultimately presented. Therefore, as an example project, the development of this research article was connected via openBIS to the necessary/received metadata and data and a clear structure was developed. The data received through openBIS as exported .zip files contain four different data types of each existing/used object type and folder, organized in a clear folder-subfolder structure. Typical standard data types used here include .doc, .txt, .json, and .html. Attached data (as Datasets) to its corresponding object type is additionally linked as a subfolder.
Acknowledgements
Author contributions
FP and SE contributed equally to this work. FP and SE prepared the initial draft of the manuscript and all authors reviewed and revised the manuscript prior to submission, as well as contributed to the conceptualizing of the paper. All authors contributed to the research and investigation process. FP and SE revised the manuscript to address recommendations offered by the anonymous Data Science Journal reviewers and all authors reviewed the revised manuscript.
Funding
This research was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) as information infrastructure (INF) project within the Collaborative Research Centre (CRC) 1411 ‘Design of Particulate Products’ (project ID: 416229255).
Conflict of interest
The authors have no competing interests to declare.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original lists citations in alphabetical order; this version lists them in order of appearance, by design. Inline URLs were turned into full citations.
- ↑ 1.0 1.1 Bauernhansl, Thomas; ten Hompel, Michael; Vogel-Heuser, Birgit, eds. (2014) (in de). Industrie 4.0 in Produktion, Automatisierung und Logistik: Anwendung · Technologien · Migration. Wiesbaden: Springer Fachmedien Wiesbaden. doi:10.1007/978-3-658-04682-8. ISBN 978-3-658-04681-1. https://link.springer.com/10.1007/978-3-658-04682-8.
- ↑ 2.0 2.1 Lasi, Heiner; Fettke, Peter; Kemper, Hans-Georg; Feld, Thomas; Hoffmann, Michael (1 August 2014). "Industry 4.0" (in en). Business & Information Systems Engineering 6 (4): 239–242. doi:10.1007/s12599-014-0334-4. ISSN 1867-0202. http://link.springer.com/10.1007/s12599-014-0334-4.
- ↑ Egger, Johannes; Masood, Tariq (1 February 2020). "Augmented reality in support of intelligent manufacturing – A systematic literature review" (in en). Computers & Industrial Engineering 140: 106195. doi:10.1016/j.cie.2019.106195. https://linkinghub.elsevier.com/retrieve/pii/S0360835219306643.
- ↑ Li, Shancang; Xu, Li Da; Zhao, Shanshan (1 April 2015). "The internet of things: a survey" (in en). Information Systems Frontiers 17 (2): 243–259. doi:10.1007/s10796-014-9492-7. ISSN 1387-3326. http://link.springer.com/10.1007/s10796-014-9492-7.
- ↑ Gisler, Michael, ed. (2001). E-Government. 1: Eine Standortbestimmung / [1. Schweizer e-Government-Symposium am 22. August 2000 in Zürich]. Michael Gisler ... (Hrsg.) (2., aktualisierte Aufl ed.). Bern Stuttgart Wien: Haupt. ISBN 978-3-258-06347-8.
- ↑ Kimmig, Julian; Zechel, Stefan; Schubert, Ulrich S. (1 February 2021). "Digital Transformation in Materials Science: A Paradigm Change in Material's Development" (in en). Advanced Materials 33 (8): 2004940. doi:10.1002/adma.202004940. ISSN 0935-9648. https://onlinelibrary.wiley.com/doi/10.1002/adma.202004940.
- ↑ Committee on Toward an Open Science Enterprise; Board on Research Data and Information; Policy and Global Affairs; National Academies of Sciences, Engineering, and Medicine (9 August 2018). Open Science by Design: Realizing a Vision for 21st Century Research. Washington, D.C.: National Academies Press. doi:10.17226/25116. ISBN 978-0-309-47624-9. https://www.nap.edu/catalog/25116.
- ↑ European Research Council Scientific Council (20 April 2022). "Open Research Data and Data Management Plans" (PDF). European Commission. https://erc.europa.eu/sites/default/files/document/file/ERC_info_document-Open_Research_Data_and_Data_Management_Plans.pdf.
- ↑ Deutsche Forschungsgemeinschaft (January 2023). "Guidelines - Collaborative Research Centres" (PDF). Deutsche Forschungsgemeinschaft. https://www.dfg.de/resource/blob/168096/3ac3b3f213ff99805d5c3d5150c5ee22/50-06-en-data.pdf.
- ↑ European Commission. Directorate General for Research and Innovation. (2016). Realising the European open science cloud: first report and recommendations of the Commission high level expert group on the European open science cloud.. LU: Publications Office. doi:10.2777/940154. https://data.europa.eu/doi/10.2777/940154.
- ↑ 11.0 11.1 11.2 Mons, Barend; Neylon, Cameron; Velterop, Jan; Dumontier, Michel; da Silva Santos, Luiz Olavo Bonino; Wilkinson, Mark D. (7 March 2017). "Cloudy, increasingly FAIR; revisiting the FAIR Data guiding principles for the European Open Science Cloud". Information Services & Use 37 (1): 49–56. doi:10.3233/ISU-170824. https://www.medra.org/servlet/aliasResolver?alias=iospress&doi=10.3233/ISU-170824.
- ↑ 12.0 12.1 Brous, Paul; Janssen, Marijn; Vilminko-Heikkinen, Riikka (2016), Scholl, Hans Jochen; Glassey, Olivier; Janssen, Marijn et al.., eds., "Coordinating Decision-Making in Data Management Activities: A Systematic Review of Data Governance Principles" (in en), Electronic Government (Cham: Springer International Publishing) 9820: 115–125, doi:10.1007/978-3-319-44421-5_9, ISBN 978-3-319-44420-8, https://link.springer.com/10.1007/978-3-319-44421-5_9. Retrieved 2024-06-04
- ↑ 13.0 13.1 Hildebrand, Knut; Gebauer, Marcus; Hinrichs, Holger et al., eds. (2011) (in de). Daten- und Informationsqualität. Wiesbaden: Vieweg+Teubner. doi:10.1007/978-3-8348-9953-8. ISBN 978-3-8348-1453-1. http://link.springer.com/10.1007/978-3-8348-9953-8.
- ↑ Ladley, John (2012). Data governance: how to design, deploy, and sustain an effective data governance program. Waltham, MA: Morgan Kaufmann. ISBN 978-0-12-415829-0.
- ↑ 15.0 15.1 Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, IJsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship" (in en). Scientific Data 3 (1): 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. https://www.nature.com/articles/sdata201618.
- ↑ Tallon, Paul P. (1 June 2013). "Corporate Governance of Big Data: Perspectives on Value, Risk, and Cost". Computer 46 (6): 32–38. doi:10.1109/MC.2013.155. ISSN 0018-9162. http://ieeexplore.ieee.org/document/6519236/.
- ↑ Besançon, Lonni; Peiffer-Smadja, Nathan; Segalas, Corentin; Jiang, Haiting; Masuzzo, Paola; Smout, Cooper; Billy, Eric; Deforet, Maxime et al. (5 June 2021). "Open science saves lives: lessons from the COVID-19 pandemic" (in en). BMC Medical Research Methodology 21 (1): 117. doi:10.1186/s12874-021-01304-y. ISSN 1471-2288. PMC PMC8179078. PMID 34090351. https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-021-01304-y.
- ↑ Tse, Edwin G.; Klug, Dana M.; Todd, Matthew H. (25 August 2020). "Open science approaches to COVID-19" (in en). F1000Research 9: 1043. doi:10.12688/f1000research.26084.1. ISSN 2046-1402. PMC PMC7590891. PMID 33145011. https://f1000research.com/articles/9-1043/v1.
- ↑ Janssen, Marijn; Charalabidis, Yannis; Zuiderwijk, Anneke (1 September 2012). "Benefits, Adoption Barriers and Myths of Open Data and Open Government" (in en). Information Systems Management 29 (4): 258–268. doi:10.1080/10580530.2012.716740. ISSN 1058-0530. http://www.tandfonline.com/doi/abs/10.1080/10580530.2012.716740.
- ↑ Crosas, Mercè (1 January 2011). "The Dataverse Network®: An Open-Source Application for Sharing, Discovering and Preserving Data" (in en). D-Lib Magazine 17 (1/2). doi:10.1045/january2011-crosas. ISSN 1082-9873. http://www.dlib.org/dlib/january11/crosas/01crosas.html.
- ↑ Lecarpentier, Damien; Wittenburg, Peter; Elbers, Willem; Michelini, Alberto; Kanso, Riam; Coveney, Peter; Baxter, Rob (14 June 2013). "EUDAT: A New Cross-Disciplinary Data Infrastructure for Science". International Journal of Digital Curation 8 (1): 279–287. doi:10.2218/ijdc.v8i1.260. ISSN 1746-8256. http://ijdc.net/article/view/8.1.279.
- ↑ Clayton, Ellen Wright; Evans, Barbara J; Hazel, James W; Rothstein, Mark A (25 October 2019). "The law of genetic privacy: applications, implications, and limitations" (in en). Journal of Law and the Biosciences 6 (1): 1–36. doi:10.1093/jlb/lsz007. ISSN 2053-9711. PMC PMC6813935. PMID 31666963. https://academic.oup.com/jlb/article/6/1/1/5489401.
- ↑ Hummel, Patrik; Braun, Matthias; Tretter, Max; Dabrock, Peter (1 January 2021). "Data sovereignty: A review" (in en). Big Data & Society 8 (1): 205395172098201. doi:10.1177/2053951720982012. ISSN 2053-9517. http://journals.sagepub.com/doi/10.1177/2053951720982012.
- ↑ 24.0 24.1 24.2 24.3 24.4 24.5 Barillari, Caterina; Ottoz, Diana S. M.; Fuentes-Serna, Juan Mariano; Ramakrishnan, Chandrasekhar; Rinn, Bernd; Rudolf, Fabian (15 February 2016). "openBIS ELN-LIMS: an open-source database for academic laboratories" (in en). Bioinformatics 32 (4): 638–640. doi:10.1093/bioinformatics/btv606. ISSN 1367-4811. https://academic.oup.com/bioinformatics/article/32/4/638/1743839.
- ↑ Bespalov, Anton; Michel, Martin C.; Steckler, Thomas, eds. (2020) (in en). Good Research Practice in Non-Clinical Pharmacology and Biomedicine. Handbook of Experimental Pharmacology. 257. Cham: Springer International Publishing. doi:10.1007/978-3-030-33656-1. ISBN 978-3-030-33655-4. https://link.springer.com/10.1007/978-3-030-33656-1.
- ↑ Machina, Hari K.; Wild, David J. (1 April 2013). "Laboratory Informatics Tools Integration Strategies for Drug Discovery: Integration of LIMS, ELN, CDS, and SDMS" (in en). SLAS Technology 18 (2): 126–136. doi:10.1177/2211068212454852. https://linkinghub.elsevier.com/retrieve/pii/S2472630322016065.
- ↑ Kraft, Angelina; Razum, Matthias; Potthoff, Jan; Porzel, Andrea; Engel, Thomas; Lange, Frank; Van den Broek, Karina; Furtado, Filipe (4 March 2016). "The RADAR Project—A Service for Research Data Archival and Publication" (in en). ISPRS International Journal of Geo-Information 5 (3): 28. doi:10.3390/ijgi5030028. ISSN 2220-9964. https://www.mdpi.com/2220-9964/5/3/28.
- ↑ "OSF". Center for Open Science. 2023. https://osf.io/.
- ↑ "CERF - Best ELN for Total Scientific Data Management". Lab-Ally, LLC. 2023. https://cerf-notebook.com/.
- ↑ "Benchling". Benchling, Inc.. 2023. https://www.benchling.com/.
- ↑ "LabFolder". Labforward GmbH. 2023. https://www.labfolder.com/.
- ↑ "Chemotion". Karlsruhe Institute of Technology. 2023. https://chemotion.net/.
- ↑ "eLabFTW". Deltablot. 2023. https://www.elabftw.net/.
- ↑ "openBIS". Scientific IT Services - ETHZ. 2023. https://openbis.ch/.
- ↑ 35.0 35.1 35.2 35.3 35.4 35.5 Bauch, Angela; Adamczyk, Izabela; Buczek, Piotr; Elmer, Franz-Josef; Enimanev, Kaloyan; Glyzewski, Pawel; Kohler, Manuel; Pylak, Tomasz et al. (1 December 2011). "openBIS: a flexible framework for managing and analyzing complex data in biology research" (in en). BMC Bioinformatics 12 (1): 468. doi:10.1186/1471-2105-12-468. ISSN 1471-2105. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-468.
- ↑ Allan, Robert J. (2009). Virtual research environments: from portals to science gateways. Chandos information professional series (1. publ ed.). Oxford: Chandos Publ. ISBN 978-1-84334-562-6.
- ↑ Candela, Leonardo; Castelli, Donatella; Pagano, Pasquale (2013). "Virtual Research Environments: An Overview and a Research Agenda" (in en). Data Science Journal 12 (0): GRDI75–GRDI81. doi:10.2481/dsj.GRDI-013. ISSN 1683-1470. http://datascience.codata.org/articles/abstract/10.2481/dsj.GRDI-013/.
- ↑ Lave, Jean; Wenger, Etienne (27 September 1991). Situated Learning: Legitimate Peripheral Participation (1 ed.). Cambridge University Press. doi:10.1017/cbo9780511815355. ISBN 978-0-521-41308-4. https://www.cambridge.org/core/product/identifier/9780511815355/type/book.
- ↑ Reimer, TF; Carusi, A (17 January 2010). Virtual Research Environment Collaborative Landscape Study. doi:10.25561/18568. http://spiral.imperial.ac.uk/handle/10044/1/18568.