Journal:Universal LIMS-based platform for the automated processing of cell-based assays
| Full article title | Universal LIMS-based platform for the automated processing of cell-based assays |
|---|---|
| Journal | Current Directions in Biomedical Engineering |
| Author(s) | Schmieder, Florian; Polk, Christoph; Gottlöber, Felix; Schöps, Patrick; Sonntag, Frank; Deuse, Ronny; Jede, Aline; Petzold, Thomas |
| Author affiliation(s) | Fraunhofer Institute for Material and Beam Technology IWS, Qualitype GmbH |
| Primary contact | Email: florian dot schmieder at iws dot fraunhofer dot de |
| Year published | 2019 |
| Volume and issue | 5(1) |
| Page(s) | 437–40 |
| DOI | 10.1515/cdbme-2019-0110 |
| ISSN | 2364-5504 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.degruyter.com/view/j/cdbme.2019.5.issue-1/cdbme-2019-0110/cdbme-2019-0110.xml |
| Download | https://www.degruyter.com/downloadpdf/j/cdbme.2019.5.issue-1/cdbme-2019-0110/cdbme-2019-0110.xml (PDF) |
Abstract
Nowadays, cell-based assays are an elementary tool for diagnostics, animal-free substance testing, and basic research. Depending on the application, the spectrum ranges from simple static cell cultures in microtiter plates to dynamic co-cultures in complex micro-physiological systems (organ-on-a-chip). Depending on the complexity of the assay, numerous working steps have to be performed and the data from different analysis systems have to be processed, combined, and documented. A universal platform has been developed for the automated handling of cell-based assays, which combines a laboratory information management system (LIMS) with a laboratory execution system (LES), a universal laboratory automation platform and established laboratory equipment. The LIMS handles the administration of all laboratory-relevant information, the planning, control, and monitoring of laboratory processes, as well as the direct and qualified processing of raw data. Using a kidney-on-a-chip system as an example, the realization of complex cell-based assays for the animal-free characterization of the toxicity of different antibiotics will be demonstrated. In the kidney-on-a-chip system, the artificial proximal tubular barrier was formed by seeding human immortalized proximal tubule cells (RPTEC) and human blood outgrowth endothelial cells (BOEC) on ThinCert membranes. Transepithelial electrical resistance (TEER) was measured daily to evaluate the barrier function of the cellular layers. Fluid handling and TEER measurements were performed using a laboratory automation platform that communicates directly with the LIMS. The LES supported laboratory assistants in executing the manual handling steps of the experiments.
Keywords: LIMS, LES, SiLA, cell based assay, lab automation
Introduction: The challenge to automate cell-based assays
Cell-based assays are nowadays widely used in drug discovery[1], personalized medicine[2][3], and basic research. This is due to the fact that cell sources like immortalized cell lines and iPS derived cells[4] offer a new range of possibilities regarding the assay sensitivity and reproducibility. Moreover, these cell sources could be harvested and cultivated as donor-specific, resulting in a wide range of patient-specific assay options. As such, many clinical researchers are developing cell-based assays to predict drug-specific interactions in vitro. In many therapeutic schemes, interactions of drugs with kidney-specific cells of the tubular and/or glomerular compartment, as well as the disruption of the kidney barriers formed by those cells, are of major interest.[5] During the past decade, many steps of assay management, including media exchange and online monitoring via imaging, have been automated using laboratory automation platforms.[6][7] Nevertheless, these assays contain a multitude of steps, including initial cell seeding, supplementation, and measurement, which are executed manually. This could lead to a multitude of problems, particularly if during all the steps data integrity is not adequately considered. Therefore, laboratory informatics is needed to translate the tools of information technology into practical science by managing electronic records to optimize laboratory operations. Multiple systems may fulfill these requirements, including laboratory information management systems (LIMS) and scientific data management systems (SDMS).[8] Based on a nephrotoxicity assay, we will show how show how using a LIMS to integrate manual and automated workflows and complex datasets of different sources solves those issues.
The nephrotoxicity assay
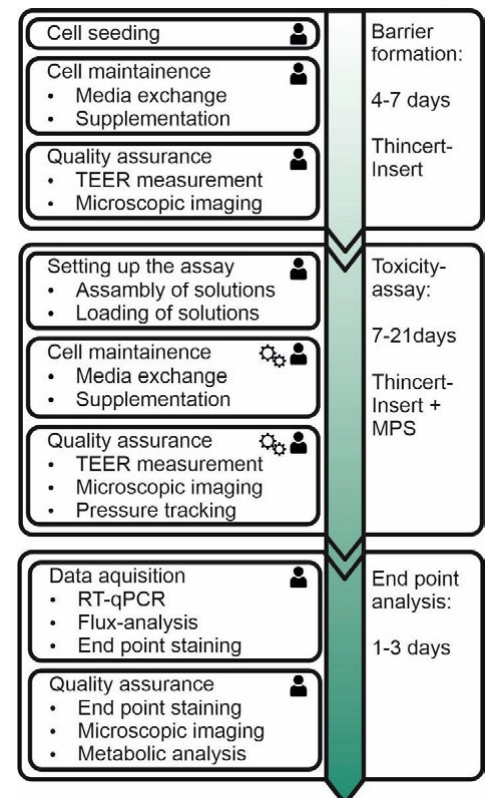
Testing nephrotoxicity in vitro means to investigate malfunctions of the glomerular or tubular barrier using cell culture models of those. Therefore the cellular assay is based on ThinCert inserts containing barrier models. Because the majority of molecular transporters—which play an important role in secretion and reabsorption of drugs—are located in the proximal tubule, most toxic effects arise in that part of the kidney.[9] As such, toxicity testing is based on an in vitro model of the proximal tubule. The whole assay consists of three main processes, with several sub-processes and steps, shown in Figure 1. It always starts with the formation of the tubular barrier. This process contains several manual sub-processes like cell seeding, cell maintenance, and quality assurance. These sub-processes are divided into working steps, e.g., "media exchange." The total time to form the barrier is about four to seven days. When the barrier is formed and characterized—to ensure quality aspects—the toxicity assay is ready to begin. Taking clinically relevant malfunctions during medication into account, several parameters including cell viability, barrier integrity, and metabolic analysis have to be investigated. To emulate in vivo physiological conditions, regarding pressure and shear stress, the ThinCert inserts are integrated into a micro-physiological system (MPS) called a ZEBRA-Chip.[10] Like during barrier formation, manual steps including assembly and loading of solutions are performed. Nevertheless, the execution of several working steps during cell maintenance and quality assurance is done by a laboratory automation platform and the MPS control unit. This means that, in contrast to barrier formation, sub-processes like media exchange and TEER measurement are run automatically. The last process of the assay is endpoint analysis. Caused by staining exposure times, this process takes one to three days and is performed manually. In general the whole assay consists of 10 sub-processes performed by laboratory assistants and three sub-processes that are performed by laboratory robots or the MPS control unit. The most crucial sub-process in laboratory automation is TEER measurement, as it is performed manually and is automatic within the same assay. Hence TEER measurement is used to show how automation and integration into a laboratory execution system (LES) is done.
|
Putting LIMS and LES to work: An example
A LIMS is a software application used by laboratories to acquire, analyze, store, and monitor laboratory data. The LIMS improves work processes by increasing productivity, traceability, sample turnaround time, and data security in many laboratories, including those in the life sciences industry.[11][12] As requirements of laboratories change, the demand for advanced data management solutions continues to grow, resulting in the development of modern, flexible LIMS capable of integrating SDMS, LES, and other informatics solutions.[11] The LIMS is the backbone of a laboratory, supporting the entire operation. Therefore, it provides all information from a single source and eliminates communication gaps between laboratory assistants, scientists, and other stakeholders, while also complying with the regulatory requirements found in the life sciences. Other benefits include more efficient laboratory workflows, simple retrieval of laboratory data, accelerated validation processes, and quality assurance of data. Beyond the LIMS, an LES can also aid in the execution of laboratory workflows, including material and product release, as well as quality testing. For laboratory professionals, an LES replaces manual data tracking with electronic processes and automates the interaction with methods, instruments, and supplies in routine laboratory procedures. By automated execution, protocols are completed efficiently and in a compliant and controlled fashion. In the life sciences, an LES is ideally suited to support key procedures in quality control and laboratory testing. Thus, method execution, calibration of instruments, data acquisition from instruments, data exchange, and data storage, as well as data review and processing can be executed through the LES.
The following subsections provide an example of combining these technologies for the automated processing of cell-based assays.
Automation platform
The developed LES is based on Qualitype GmbH's Abetter LIMS platform. A two-level portal robotics platform (PRP) manufactured by GeSiM mbH was used for laboratory automation, specifically in regards to the handling of liquids, imaging, and the measurement of TEER.[6] Microtiter plates and kidney-on-a-chip systems were placed on a temperature-controlled holder plate located between the two bridges. A versatile tool handler was mounted to the upper bridge, which can also be equipped with various fluid handling systems, sensors (TEER-Electrode) and tools. Another tool handler was mounted to the lower bridge. Each well or kidney-on-a-chip system could then be maintained separately with different cell culture media as well as monitored by fluorescence imaging and TEER-measurement.
LIMS integration via LES
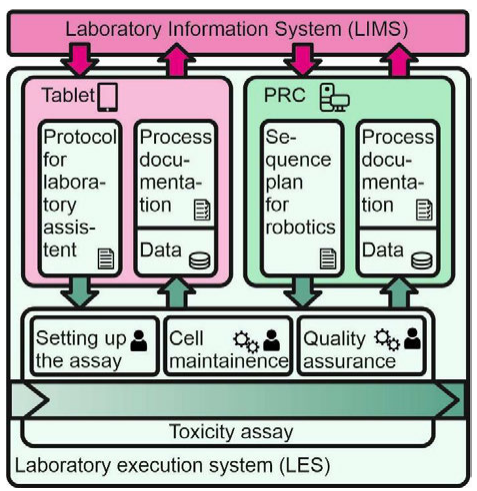
To make use of the beneficial effects that are described above for automation of data handling and processing of the nephrotoxicity assay, a universal LIMS platform was developed. It combines a LIMS with a LES for the automated handling of cell-based assays, while also integrating a universal laboratory automation platform and established laboratory equipment. Figure 2 shows the management of all laboratory-relevant information within the described assay monitored by the LIMS, such as the planning, control, and monitoring of laboratory processes, as well as the direct and qualified processing of raw data. Thus, it induces the work steps according to the assay's protocol via the LES to the laboratory assistant or via portal robotic control (PRC) to the PRP. The LES guides the laboratory assistant through the manual work steps of the experiments by using an interactive graphical user interface. The data gained during the experiment, as well as the documentation, recorded by checkboxes in-between work steps by the LES, are returned to the LIMS.
|
SiLA - Standardization in Lab Automation
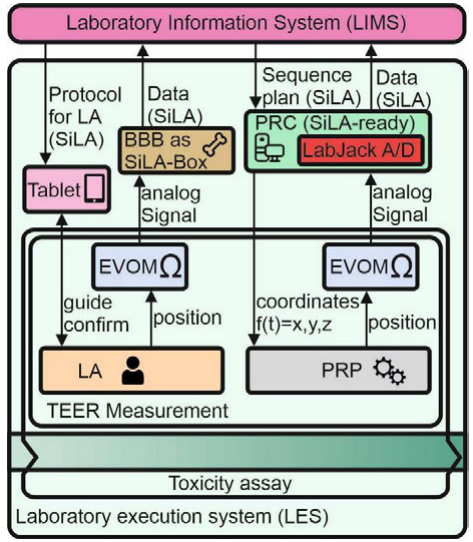
Conforming with the data management procedures shown in Figure 2, a powerful but universal data management standard is needed to face the obstacles during integration and implementation of laboratory informatics solutions.[12] Associations such as SiLA (Standardization in Lab Automation) and the International Consortium for Innovation and Quality in Pharmaceutical Development (IQ Consortium) are making significant attempts to introduce new interfaces and data management standards to facilitate system integration.[13] At the moment, the lack of integration standards is a major challenge to the greater application of laboratory informatics in many laboratories. With no unified strategy, laboratories largely continue to follow conventional procedures, regardless of efficiency, connectivity and interoperability between systems. SiLA standards enable a modular and versatile system approach to solve this issue. In line with these efforts, the TEER Measurement System was integrated as shown in Figure 3.
|
For manual handling of TEER measurement, the laboratory assistant is guided by the LES. The TEER measurement device (EVOM) was connected to a BeagleBone black (BBB) that converts analog to digital signals and acts as a data converter to the SiLA standard. Afterwards, data are transferred to the LIMS. Automated processes by the PRP are controlled by a SiLA-ready computer that acts as as a PRC and integrates a LabJack A/D converter for transformation of analog signals.
Conclusion
This work shows how a combination of LIMS and LES can be used to integrate various manual and automated laboratory processes of cell-based assays within one informatics solution. Standard analog laboratory devices, like the TEER system used here, can be integrated easily in such systems by using embedded computers or hardware that can already function as a SiLA converter. The combination of laboratory informatics, devices, and universal gateways therefore helps to improve quality of acquired data and reduces sample turnaround time.
Acknowledgements
Funding
This study was funded by the European Union (SAB project 100216519 – CeCuLab) and the Federal Ministry of Economics and Technology (BMWi) according to a decision of the German Federal Parliament (IGF project 19175 BR/1 - ZEBRA).
Conflict of interest
Ronny Deuse, Aline Jede, and Thomas Petzold work at Qualitype GmbH, which develops and sells laboratory information management systems. All other authors state no conflict of interest.
References
- ↑ Hering, Y.; Berthier, A.; Duez, H. et al. (2018). "Development and implementation of a cell-based assay to discover agonists of the nuclear receptor REV-ERBα". Journal of Biological Methods 5 (3): e94. doi:10.14440/jbm.2018.244. PMC PMC6706147. PMID 31453244. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6706147.
- ↑ Blom, K.; Nygren, P.; Alvarsson, J. et al. (2016). "Ex Vivo Assessment of Drug Activity in Patient Tumor Cells as a Basis for Tailored Cancer Therapy". Journal of Laboratory Automation 21 (1): 178–87. doi:10.1177/2211068215598117. PMID 26246423.
- ↑ Blom, K.; Nygren, P.; Larsson, R. et al. (2017). "Predictive Value of Ex Vivo Chemosensitivity Assays for Individualized Cancer Chemotherapy: A Meta-Analysis". SLAS Technology 22 (3): 306-314. doi:10.1177/2472630316686297. PMID 28378608.
- ↑ Rowe, R.G.; Daley, G.Q. (2019). "Induced pluripotent stem cells in disease modelling and drug discovery". Nature Reviews Genetics 20 (7): 377-388. doi:10.1038/s41576-019-0100-z. PMC PMC6584039. PMID 30737492. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6584039.
- ↑ Schmieder, F.; Sradnick, J.; Hempel, M. et al. (28 September 2018). "Emulating gentamycin induced nephrotoxicity in an artificial system of the renal proximal tubule". ResearchGate. Fraunhofer Institute for Material and Beam Technology. doi:10.13140/RG.2.2.12274.96960. https://www.researchgate.net/publication/328172169_Emulating_gentamycin_induced_nephrotoxicity_in_an_artificial_system_of_the_renal_proximal_tubule.
- ↑ 6.0 6.1 Schmieder, F.; Polk, C.; Eger, R. et al. (2014). "Robotic platform for the automated handling and monitoring of cell culture devices". Proceedings of Bionection 2014. doi:10.13140/RG.2.1.2468.4247.
- ↑ Prabhu, G.R.D.; Urban, P.L. (2017). "The dawn of unmanned analytical laboratories". TrAC Trends in Analytical Chemistry 88: 41–52. doi:10.1016/j.trac.2016.12.011.
- ↑ Neubert, S.; Göde, B.; Gu, X. et al. (2017). "Potential of Laboratory Execution Systems (LESs) to Simplify the Application of Business Process Management Systems (BPMSs) in Laboratory Automation". SLAS Technology 22 (2): 206-216. doi:10.1177/2211068216680331. PMID 27908978.
- ↑ Yin, J.; Wang, J. (2016). "Renal drug transporters and their significance in drug-drug interactions". Acta Pharmaceutica Sinica B 6 (5): 363-373. doi:10.1016/j.apsb.2016.07.013. PMC PMC5045553. PMID 27709005. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5045553.
- ↑ Schmieder, F.; Förster, D.; Hempel, M. et al. (23 May 2018). "Mimicking human physiology at Transwell-based barrier models of the proximal tubule – The ZEBRA-Chip". ResearchGate. Fraunhofer Institute for Material and Beam Technology. doi:10.13140/RG.2.2.27364.48002. https://www.researchgate.net/publication/325398524_Mimicking_human_physiology_at_Transwell-based_barrier_models_of_the_proximal_tubule_-_The_ZEBRA-Chip.
- ↑ 11.0 11.1 Tagger, B. (20 February 2026). "An Introduction and Guide to Successfully Implementing a LIMS (Laboratory Information Management System)" (PDF). Computer Science Department, University of Wales. http://www0.cs.ucl.ac.uk/staff/B.Tagger/LimsPaper.pdf. Retrieved 05 June 2019.
- ↑ 12.0 12.1 Paszko, C.; Turner, E. (2001). Laboratory Information Management Systems (2nd ed.). CRC Press. ISBN 9780824741412. http://books.google.com/books?id=ycEqnzPl2lYC.
- ↑ Bär, H.; Hochstrasser, R.; Papenfub, B. (2012). "SiLA: Basic standards for rapid integration in laboratory automation". Journal of Laboratory Automation 17 (2): 86–95. doi:10.1177/2211068211424550. PMID 22357556.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. We also added PMCID and DOI when they were missing from the original reference.