Journal:Chemozart: A web-based 3D molecular structure editor and visualizer platform
| Full article title | Chemozart: A web-based 3D molecular structure editor and visualizer platform |
|---|---|
| Journal | Journal of Cheminformatics |
| Author(s) | Mohebifar, Mohamad; Sajadi, Fatemehsadat |
| Author affiliation(s) | Shahid Beheshti University |
| Primary contact | Email: mohamad@mohebifar.com |
| Year published | 2015 |
| Volume and issue | 7 |
| Page(s) | 56 |
| DOI | 10.1186/s13321-015-0101-7 |
| ISSN | 1758-2946 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://jcheminf.springeropen.com/articles/10.1186/s13321-015-0101-7 |
| Download | http://jcheminf.springeropen.com/track/pdf/10.1186/s13321-015-0101-7 (PDF) |
Abstract
Background: Chemozart is a 3D Molecule editor and visualizer built on top of native web components. It offers an easy to access service, user-friendly graphical interface and modular design. It is a client centric web application which communicates with the server via a representational state transfer style web service. Both client-side and server-side application are written in JavaScript. A combination of JavaScript and HTML is used to draw three-dimensional structures of molecules.
Results: With the help of WebGL, three-dimensional visualization tool is provided. Using CSS3 and HTML5, a user-friendly interface is composed. More than 30 packages are used to compose this application which adds enough flexibility to it to be extended. Molecule structures can be drawn on all types of platforms and is compatible with mobile devices. No installation is required in order to use this application and it can be accessed through the internet. This application can be extended on both server-side and client-side by implementing modules in JavaScript. Molecular compounds are drawn on the HTML5 Canvas element using WebGL context.
Conclusions: Chemozart is a chemical platform which is powerful, flexible, and easy to access. It provides an online web-based tool used for chemical visualization along with result oriented optimization for cloud based API (application programming interface). JavaScript libraries which allow creation of web pages containing interactive three-dimensional molecular structures has also been made available. The application has been released under Apache 2 License and is available from the project website https://chemozart.com.
Keywords: Cheminformatics, web-based, JavaScript, visualization
Background
In the field of computational chemistry, applications which are capable of constructing and viewing 3D structures of molecules play an important role. Such software can be used by students to understand stereochemical concepts.[1] There are numerous desktop applications available for viewing and building 3D molecules. Avogadro[2], Jmol[3], QuteMol[4] and PyMol are few such examples. When it comes to web applications capable of constructing 3D chemical structures, there are not many available.
Today web-based tools are becoming extremely popular. There are numerous benefits which can be derived from them; accessibility, flexible core technologies, platform independence and compatibility are some of them. Most of the web applications which are used for building chemical structures have limited capabilities and most of them are two-dimensional editors such as ChemDoodle.[5] There are several chemical structure editors available powered by Java applets such as JME, a free 2D molecule editor java applet[6]; however, they are not compatible with all the browsers. Besides, Java needs to be separately installed on the system in order to run these applications. This issue exists for other embedded objects like Flash and Flame[7], a Flash molecular editor is a case in point. There is another web application available to build 3D chemical structures, it is called CH5M3D.[8] It portrays a 3D picture with HTML5; however, it fails to deliver optimum results as it uses canvas 2D context. Because of its limitation, it does not use any shader program. As a result, three-dimensional rendering cannot be portrayed in the truest sense. Today, it is possible to draw sophisticated graphics that are hardware accelerated by the GPU thanks to HTML5 and WebGL. Furthermore, these components are supported by all modern web browsers, especially mobile browsers.
Applications built using web-based components can be accessed with ease on portable devices such as iPods and smartphones with an additional feature which enables use of it in offline mode too. In today's tech-savvy world, the utilization of chemistry-related applications is dramatically growing.[9] This 3D molecule editor targets students at varied levels of study, i.e. high school, college, and graduate school. Along with the students Chemozart targets, chemical professionals and teachers also can effectively research in any topic and appropriately solve the queries of their students. We have developed a 3D molecule editor on top of web components which leads to better performance and maintainability. With the help of node.js (https://nodejs.org/), it is now possible to use JavaScript outside the web browsers. Both server-side and client-side codes are fully written in JavaScript. By writing both in the same language, we seek to enhance integration. JavaScript is the language used to develop this software. A Github data visualization factually states that JavaScript is considered to have the most active repositories. As a result, there are diverse packages written in JavaScript with which this software can be bundled.
A chemical toolbox is also needed in order to read different chemical file formats, calculate energy, etc. OpenBabel is considered to be one of the best open-source chemical toolkits, and it can be easily ported to different languages. There are bindings of OpenBabel in some languages such as Rubabel[10] for Ruby and Pybel[11] for Python. We also made OpenBabel-Node (http://mohebifar.github.io/OpenBabel-Node/) to port OpenBabel[12] to node.js. It exposes OpenBabel application programming interfaces (APIs) to many available packages in node.js via a convenient interface. We bundled it with express.js web framework to create a chemical representational state transfer style RESTful API. OpenBabel-Node is used to read and write a variety of chemical file formats. Apart from this, it also supports various molecular mechanics force fields and provides optimization of geometry of the molecules.
Chemozart is a web application tool that can be used for viewing and editing of 3D molecular structures. With the help of this web-based platform, users can easily create, modify, or view the structures of the molecular compounds. With the help of JavaScript and HTML, users can easily draw or view the 3D structures of the molecular compounds. The web application represents the molecular structures in both of the client-side and server-side applications. The client-side consists of the UI and the visualization part that helps one to view or edit the structures easily. The convenient user interface allows for modifying molecular structures interactively, while the visualization part helps in viewing the 3D representations of the molecular compounds.
Implementation
Software architecture and interactivity
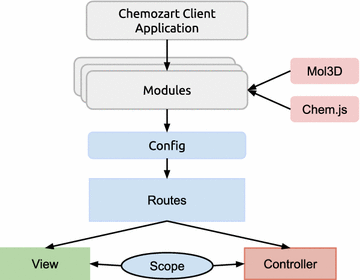
Chemozart provides a web-based platform for creation, modification and display of molecules. It is available under the Apache 2 License, which gives power and access to everyone to contribute to and extend it. This package consists of a client-side and a server-side application. The client-side application is designed according to the Model View ViewModel (MVVM) pattern. It is built with Angular.js which is an MV* client-side framework. It offers service providers, two-way bindings, data models, convenient RESTful resource client service, declarative user interface, etc. The server-side application is designed according to the Model View Controller (MVC) pattern. It is also completely written in JavaScript and uses node.js as the runtime environment. This package is flexible enough to be developed with a well-organized structure. The entire application is written in JavaScript according to ECMAScript 6, which offers classy, flexible, and cleaner syntax, a universal way for module definition, etc. Chemozart transpiles ES6 codes to ES5-friendly codes using Babel (https://babeljs.io/) for browser support. The stylesheets are originally written in LESS, which are compiled to CSS.
The client-side application depends on two major packages, which are designed to facilitate the process of binding chemical data to the graphics. Chem.js is a JavaScript package which creates event-based models of molecules, atoms, and bonds through an object-oriented approach. It is also used to serialize the molecules in JSON format. Mol3D (https://github.com/mohebifar/mol3d) is another JavaScript package that displays the structure of molecules, and it uses Three.js to render 3D objects with WebGL. It converts the Chem.js objects into Three.js (https://github.com/mrdoob/three.js) objects according to the atoms’ position, atomic radius, and CPK coloring convention. The default display mode is “ball and stick” as it is the most convenient model for editing molecules. It is also possible to make arbitrary display modes.
The server-side application depends on OpenBabel-Node, which is a sub-module of Chemozart, to provide native bindings of OpenBabel in node.js. It is originally written in C++ and makes it possible to use OpenBabel APIs in JavaScript code. This package is used to build 3D coordinates, calculate the energy of molecules, and import and export chemical files, including SMILES[13], CML[14], Hyperchem HIN, MDL SD[15], Sybyl Mol2[16] and PDB.[17]
One of the most important aims of Chemozart is to provide a good user experience. So it facilitates building molecules with the help of mouse and keyboard actions or by touch screen devices. Changing the position of atoms is also as simple as dragging them around. All the hidden elements such as the periodic table appear with an animation effect that provides a better user experience.
While this application focuses on educational purposes, more educational functionalities are planned to be included in future versions. The most important feature is to attach a database to the server-side application to make it possible to share a molecule via a link so that students can share their thoughts and questions with their teachers. It also facilitates the process of teaching for teachers given that this application works on mobile devices. In addition, more OpenBabel features will be used such as force field clean-up. More complete support for chemical file formats is also planned to be included in future versions (Fig. 1).
|
Installation
One of the unique features of this application is the fact that it does not require an installation process and can be accessed online. However, it can be installed locally too. The application requires an existing installation of node.js, npm, bower, and grunt. The code is developed and maintained on a Git repository (https://github.com/mohebifar/chemozart) available on Github. After cloning this repository in any convenient location, dependencies are required to be installed. Note that the client-side dependencies are managed by bower and the server-side dependencies are managed by npm. It is required to run “npm install” and “bower install” commands in a terminal window to install both frontend and backend dependencies. By running “grunt serve” the application will be started and it starts listening on port 9000 by default while a web browser will show up automatically. It is also possible to change the port by changing the OS environment variable “PORT”.
This application requires a web server since it has some functions that use OpenBabel-node. However, it can also be accessed without web server installation, but some functionalities that require OpenBabel will not be available such as energy calculation, adding Hydrogens, and the 3D build, export, and import of different chemical languages. The different versions of this application are distributed as compressed zip archives and are accessible in the releases menu on the Github page.
Implementation
Library development
The client-side consists of the user interface along with a variety of logical modules. It helps to view or edit the structures interactively. The user interface is composed in CSS and HTML, and a dynamic stylesheet language called LESS is used in this software. These stylesheets are compiled into CSS, which makes it easier to write and maintain stylesheets in big projects. For the views, JADE platform is used, offering inheritance and re-usable functions. Based on the MVVM architectural pattern, the molecular models are two-way bound as ViewModel between the controllers and view. It exposes the data objects in such a way that the user can view the model structure without being bothered about the back-end logic of the model. Working under a framework of Angular.js, which is based on MV* architecture, the application firstly reads the view and interprets it as directives and binds the data to a model. The models here are a representation of a molecule drawn or structured by the working user. Further, the models discussed above are originally the atoms and bonds.
The server-side of the application is based on the MVC software architectural pattern, which helps to separate this web application into three interconnected parts. This helps in easy viewing of the structure of a molecular compound by separating the inner information from the logics and views. Written in JavaScript, the software uses both node.js and express.js, which can be used for fast running of this application with fewer lines of codes to develop.
Modes
The 3D molecule editor and visualizer works on three modes which are as follows:
- Camera mode
- Editing mode
- Positioning mode
Camera mode
In this mode the user can rotate any molecular structure just by holding the left button of the mouse and moving it. Its function Pan can also be used just by holding the right button of the mouse and moving it according to requirements. The zoom function of this mode can be accessed when the user scrolls by holding the middle button of the mouse. This mode works with touch screen devices as well.
Editing mode
Editing any molecular structure is quite easy because of the array of options available in this mode. To add an atom to any structure the user has to click on the empty space, while to delete any atom they just right click on the atom itself. By dragging from one atom to another, the user can add a bond. Clicking on a bond adjusts their order. Removing a bond is also possible by right clicking on it.
Positioning mode
This mode offers proper positioning of the molecular structure. In this mode, dragging the atoms on the screen changes the position of them.
Menus
The menu option of this chemical structure viewing software comprises of three main options which are known as File, Build, and Energy.
File
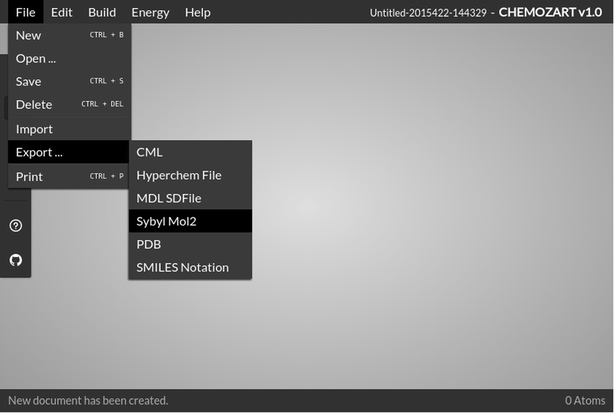
The file option of this web application comprises an array of options which includes New, Open, Save, Delete, Import, Export, and Print. The user can save or delete a structure easily or even import one from the desktop with the help of these options available in the File menu. It supports a wide range of file formats to read and write. All the drawn structures are also saved in the browser’s local storage.
Build
The Build menu comprises of two options which are known as Build 3D and Add Hydrogens. The Build 3D option helps user to generate 3D coordinates for the drawn molecule. Adding Hydrogen functionality to the existing structure is also provided in order to fill out implicit valence spots. The Builder and Hydrogen Adder options use OpenBabel-Node via a RESTful API.
Energy
Different molecular mechanics force-field methods like MMFF94, UFF, and Ghemical are available to evaluate the energy. By creating the structure and using any of these options, the energy shows up in a dialog. Further, this is a common feature of the OpenBabel-Node too.
Results and discussion
Technologies and components
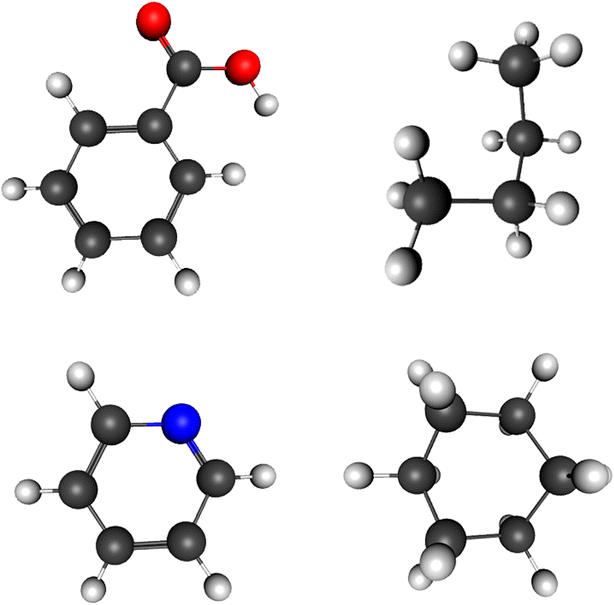
This web application tool is used for viewing and editing of molecular structures as well as to view them in 3D mode. One can easily draw any structure with the help of JavaScript codes and represent it on the HTML5 canvas element with WebGL context. By using standard techniques, the 3D structure of a molecule can also be viewed. Because of its flexibility in operation and numerous editing and modifying modes, the software is easy to work with and beneficial. With its numerous editing options and an easy way to construct or view structures, this chemical software is good for educating and explaining the detailed aspects of 3D geometry in a convenient way. This 3D geometry visualization also helps in performing an in depth analysis of the background of the image and getting clear information about the formation of each molecule, which is not possible with 2D images with no visual of background images. Analysis of molecular structures and their 3D representations also play an important role in the field of computational chemistry as well. The application does not require any installation and can be accessed online. Interested users can install it locally and continue work without any hassle. It provides a good user experience with the help of keyboard and mouse actions as well as with touch screen devices. In accordance with the educational requisites, Chemozart will give students clarity about molecules and their stereochemistry concepts. The biggest advantage of this software is that it assists in educating the students and imparting knowledge to them as it works on portable devices (Fig. 2).
|
This application is written in JavaScript, which is the preferred language for making web applications. Usage of JavaScript enhances the integration as it is used on both the client-side and server-side. Moreover, it helps in creating an easy user interface, and any one can avail its advanced functionalities just by entering a web url in the browser. Now studying a 3D structure of a molecular compound, modifying it, or calculating the energy is easier because of this online chemical software. Chemozart is surely going to be a milestone in the construction of 3D images in the field of chemistry to analyze, study, and create unique 3D chemical structure images with its user-friendly interface.
Creating web pages
This application can be accessed in two different environments: development and production. To create a web page, development files should be used, which make it easier to develop and extend the application. After changing the code files, by running “grunt build,” production files will be created in a folder named “dist”. It basically concatenates and compresses the scripts and stylesheets to prevent network delay caused by transferring all unnecessary characters in the code such as white spaces. To create a chemical editor web page using Chemozart, it is required to verify that the angular.js that the web page belongs to is a Chemozart application. This can be accomplished by adding the following line to the <body> statement:
<body ng-app = “chemartApp”>
The main element to create a Chemozart web page is the div element with a ui-view attribute. It will automatically include the tool bar, status bar, menu, and the drawing window to show and edit the molecule. It is accomplished by using angular.js templates.
<div ui-view = “”> </div>
At the end of the body tag, it is required to include all the dependencies in the right order that vary from vendor libraries to each Chemozart modules.
Each template can be customized as well as the stylesheets. Templates in JADE and their corresponding stylesheets in LESS are located inside the app or components folder inside the client folder based on their role. Components such as menu bar, tool bar, status bar, periodic table, help window, and about window are located in the components folder and the main template that puts these components together is located in the app folder (Fig. 3).
|
User-defined functions
This application is built with Angular.js on client-side. All the components and libraries are a directive, a service, or a controller. For example, to create a button in the menu bar that adds a carbon on the drawing window, the following steps could be used. In the first step, a service must be created.
This service returns a function that creates an instance of “Chem.Atom” that is a class of mol.js module. Then the atomic number should be determined by assigning the value to the “atomicNumber” property. To “position” property determines the atom’s position that should be an instance of “Three Vector”. A singleton of the canvas object can be accessed via a service named “canvas”. To add the creation to the drawing window, the atom should be passed to the “addAtom” method on the canvas object. This service should be injected to the main controller (client/app/main/main.controller.js) by adding “addCarbon” to the list of arguments of the main controller function. After injecting the service, it should be assigned to the controller’s scope so it can be accessed in the view.
Finally, a button in the view is needed. To add this button in the menu bar view, the JADE file of the menu bar (client/components/menubar/menubar.jade) should be altered as following:
Conclusions
It can be concluded that Chemozart is a chemical web-based application and component which provides the ability to create 3D structures of molecules. The most important feature is that there’s no need to install anything and it can be accessed easily via a URL. Also, it can be beneficial for educational purposes as well. To catch up with the technology, Chemozart has been designed in a way that it is compatible with mobile devices. The most up-to-date version of this application is available on the project home page. For the purpose of smooth functioning, it is advisable to access Chemozart using the latest versions of browsers across different operating systems (Fig. 4)
|
Availability and requirements
Project name: Chemozart
Project home page: https://chemozart.com
Operating system(s): Platform independent
Programming language: JavaScript
Other requirements: An up-to-date web browser
License: Apache2 License
Any restrictions to use by non-academics: None
Declarations
Authors’ contributions
MM developed the applications and drafted the manuscript. FS participated in the design of study, helped to write Chem.js and Mol3D and helped to draft the manuscript. Both authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to Shant Shahbazian for reading a previous draft of this paper and some helpful suggestions.
Competing interests
The authors declare that they have no competing interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
References
- ↑ Abraham, M.; Varghese, V.; Tang, H. (2010). "Using molecular representations to aid student understanding of stereochemical concepts". Journal of Chemical Education 87 (12): 1425–1429. doi:10.1021/ed100497f.
- ↑ Hanwell, M.D.; Curtis, D.E.; Lonie, D.C. et al. (2012). "Avogadro: An advanced semantic chemical editor, visualization, and analysis platform". Journal of Cheminformatics 4: 17. doi:10.1186/1758-2946-4-17. PMC PMC3542060. PMID 22889332. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3542060.
- ↑ "Jmol: An open-source Java viewer for chemical structures in 3D". SourceForge.net. Slashdot Media. http://jmol.sourceforge.net/. Retrieved 01 November 2015.
- ↑ Tarini, M.; Cignoni, P.; Montani, C. (2006). "Ambient occlusion and edge cueing for enhancing real time molecular visualization". IEEE Transactions on Visualization and Computer Graphics 12 (5): 1237–1244. doi:10.1109/TVCG.2006.115.
- ↑ Burger, M.C. (2015). "ChemDoodle Web Components: HTML5 toolkit for chemical graphics, interfaces, and informatics". Journal of Cheminformatics 7: 35. doi:10.1186/s13321-015-0085-3. PMC PMC4503857. PMID 26185528. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4503857.
- ↑ Ertl, P. (2010). "Molecular structure input on the web". Journal of Cheminformatics 2: 1. doi:10.1186/1758-2946-2-1. PMC PMC2827360. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2827360.
- ↑ Dallakian, P.; Haider, N. (2011). "FlaME: Flash Molecular Editor - A 2D structure input tool for the web". Journal of Cheminformatics 3: 6. doi:10.1186/1758-2946-3-6. PMC PMC3037917. PMID 21284863. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3037917.
- ↑ Earley, C.W. (2013). "CH5M3D: An HTML5 program for creating 3D molecular structures". Journal of Cheminformatics 5: 46. doi:10.1186/1758-2946-5-46. PMC PMC4177146. PMID 24246004. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4177146.
- ↑ Libman, D.; Huang, L. (2013). "Chemistry on the go: Review of chemistry apps on smartphones". Journal of Chemical Education 90 (3): 320–325. doi:10.1021/ed300329e.
- ↑ Smith, R.; Williamson, R.; Ventura, D.; Prince, J.T. (2013). "Rubabel: Wrapping open Babel with Ruby". Journal of Cheminformatics 5: 35. doi:10.1186/1758-2946-5-35. PMC PMC3733738. PMID 23883475. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3733738.
- ↑ O'Boyle, N.M.; Morley, C.; Hutchison, G.R. (2008). "Pybel: A Python wrapper for the OpenBabel cheminformatics toolkit". Chemistry Center Journal 2: 5. doi:10.1186/1752-153X-2-5. PMC PMC2270842. PMID 18328109. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2270842.
- ↑ O'Boyle, N.M.; Banck, M.; James, C.A. (2011). "Open Babel: An open chemical toolbox". Journal of Cheminformatics 3: 33. doi:10.1186/1758-2946-3-33. PMC PMC3198950. PMID 21982300. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3198950.
- ↑ Weininger, D. (1988). "SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules". Journal of Chemical Information and Modeling 28 (1): 31–36. doi:10.1021/ci00057a005.
- ↑ Murray-Rust, P.; Rzepa, H.S. (2003). "Chemical markup, XML, and the World Wide Web. 4. CML schema". Journal of Chemical Information and Modeling 43 (3): 757–772. doi:10.1021/ci0256541.
- ↑ Dalby, A.; Nourse, J.G.; Hounshell, W.D. et al. (1992). "Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited". Journal of Chemical Information and Modeling 32 (3): 244–255. doi:10.1021/ci00007a012.
- ↑ Homer, R.W.; Swanson, J.; Jilek, R.J. et al. (2008). "SYBYL line notation (SLN): A single notation to represent chemical structures, queries, reactions, and virtual libraries". Journal of Chemical Information and Modeling 48 (12): 2294–2307. doi:10.1021/ci7004687.
- ↑ Berman, H.M. (2008). "The Protein Data Bank: A historical perspective". Acta Crystallographica Section A 64 (Pt 1): 88–95. doi:10.1107/S0108767307035623. PMID 18156675.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Numerous grammar errors were also corrected.