Biomedical text mining
Biomedical text mining (including biomedical natural language processing or BioNLP) refers to the methods and study of how text mining may be applied to texts and literature of the biomedical domain. As a field of research, biomedical text mining incorporates ideas from natural language processing, bioinformatics, medical informatics and computational linguistics. The strategies in this field have been applied to the biomedical literature available through services such as PubMed.
In recent years, the scientific literature has shifted to electronic publishing but the volume of information available can be overwhelming. This revolution of publishing has caused a high demand for text mining techniques. Text mining offers information retrieval (IR) and entity recognition (ER).[1] IR allows the retrieval of relevant papers according to the topic of interest, e.g. through PubMed. ER is practiced when certain biological terms are recognized (e.g. proteins or genes) for further processing.
Considerations
Applying text mining approaches to biomedical text requires specific considerations common to the domain.
Availability of annotated text data

Large annotated corpora used in the development and training of general purpose text mining methods (e.g., sets of movie dialogue,[3] product reviews,[4] or Wikipedia article text) are not specific for biomedical language. While they may provide evidence of general text properties such as parts of speech, they rarely contain concepts of interest to biologists or clinicians. Development of new methods to identify features specific to biomedical documents therefore requires assembly of specialized corpora.[5] Resources designed to aid in building new biomedical text mining methods have been developed through the Informatics for Integrating Biology and the Bedside (i2b2) challenges[6][7][8] and biomedical informatics researchers.[9][10] Text mining researchers frequently combine these corpora with the controlled vocabularies and ontologies available through the National Library of Medicine's Unified Medical Language System (UMLS) and Medical Subject Headings (MeSH).
Machine learning-based methods often require very large data sets as training data to build useful models.[11] Manual annotation of large text corpora is not realistically possible. Training data may therefore be products of weak supervision[12][13] or purely statistical methods.
Data structure variation
Like other text documents, biomedical documents contain unstructured data.[14] Research publications follow different formats, contain different types of information, and are interspersed with figures, tables, and other non-text content. Both unstructured text and semi-structured document elements, such as tables, may contain important information that should be text mined.[15] Clinical documents may vary in structure and language between departments and locations. Other types of biomedical text, such as drug labels,[16] may follow general structural guidelines but lack further details.
Uncertainty
Biomedical literature contains statements about observations that may not be statements of fact. This text may express uncertainty or skepticism about claims. Without specific adaptations, text mining approaches designed to identify claims within text may mis-characterize these "hedged" statements as facts.[17]
Supporting clinical needs
Biomedical text mining applications developed for clinical use should ideally reflect the needs and demands of clinicians.[5] This is a concern in environments where clinical decision support is expected to be informative and accurate. A comprehensive overview of the development and uptake of NLP methods applied to free-text clinical notes related to chronic diseases is presented in.[18]
Interoperability with clinical systems
New text mining systems must work with existing standards, electronic medical records, and databases.[5] Methods for interfacing with clinical systems such as LOINC have been developed[19] but require extensive organizational effort to implement and maintain.[20][21]
Patient privacy
Text mining systems operating with private medical data must respect its security and ensure it is rendered anonymous where appropriate.[22][23][24]
Processes
Specific sub tasks are of particular concern when processing biomedical text.[14]
Named entity recognition
Developments in biomedical text mining have incorporated identification of biological entities with named entity recognition, or NER. Names and identifiers for biomolecules such as proteins and genes,[25] chemical compounds and drugs,[26] and disease names[27] have all been used as entities. Most entity recognition methods are supported by pre-defined linguistic features or vocabularies, though methods incorporating deep learning and word embeddings have also been successful at biomedical NER.[28][29]
Document classification and clustering
Biomedical documents may be classified or clustered based on their contents and topics. In classification, document categories are specified manually,[30] while in clustering, documents form algorithm-dependent, distinct groups.[31] These two tasks are representative of supervised and unsupervised methods, respectively, yet the goal of both is to produce subsets of documents based on their distinguishing features. Methods for biomedical document clustering have relied upon k-means clustering.[31]
Relationship discovery
Biomedical documents describe connections between concepts, whether they are interactions between biomolecules, events occurring subsequently over time (i.e., temporal relationships), or causal relationships. Text mining methods may perform relation discovery to identify these connections, often in concert with named entity recognition.[32]
Hedge cue detection
The challenge of identifying uncertain or "hedged" statements has been addressed through hedge cue detection in biomedical literature.[17]
Claim detection
Multiple researchers have developed methods to identify specific scientific claims from literature.[33][34] In practice, this process involves both isolating phrases and sentences denoting the core arguments made by the authors of a document (a process known as argument mining, employing tools used in fields such as political science) and comparing claims to find potential contradictions between them.[34]
Information extraction
Information extraction, or IE, is the process of automatically identifying structured information from unstructured or partially structured text. IE processes can involve several or all of the above activities, including named entity recognition, relationship discovery, and document classification, with the overall goal of translating text to a more structured form, such as the contents of a template or knowledge base. In the biomedical domain, IE is used to generate links between concepts described in text, such as gene A inhibits gene B and gene C is involved in disease G.[35] Biomedical knowledge bases containing this type of information are generally products of extensive manual curation, so replacement of manual efforts with automated methods remains a compelling area of research.[36][37]
Information retrieval and question answering
Biomedical text mining supports applications for identifying documents and concepts matching search queries. Search engines such as PubMed search allow users to query literature databases with words or phrases present in document contents, metadata, or indices such as MeSH. Similar approaches may be used for medical literature retrieval. For more fine-grained results, some applications permit users to search with natural language queries and identify specific biomedical relationships.[38]
On 16 March 2020, the National Library of Medicine and others launched the COVID-19 Open Research Dataset (CORD-19) to enable text mining of the current literature on the novel virus. The dataset is hosted by the Semantic Scholar project[39] of the Allen Institute for AI.[40] Other participants include Google, Microsoft Research, the Center for Security and Emerging Technology, and the Chan Zuckerberg Initiative.[41]
Resources
Corpora
The following table lists a selection of biomedical text corpora and their contents. These items include annotated corpora, sources of biomedical research literature, and resources frequently used as vocabulary and/or ontology references, such as MeSH. Items marked "Yes" under "Freely Available" can be downloaded from a publicly accessible location.
| Corpus Name | Authors or Group | Contents | Freely Available | Ref. |
|---|---|---|---|---|
| 2019 Bacteria Biotope | BioNLP-OST | Annotated scientific and textbook texts to recognize mentions of microorganisms, microbial biotopes and phenotypes, to normalize these mentions according to the knowledge resources of the field, and to extract the relationships between them. | Yes | [42] |
| 2006 i2b2 Deidentification and Smoking Challenge | i2b2 | 889 de-identified medical discharge summaries annotated for patient identification and smoking status features. | Yes, with registration | [43][44] |
| 2008 i2b2 Obesity Challenge | i2b2 | 1,237 de-identified medical discharge summaries annotated for presence or absence of comorbidities of obesity. | Yes, with registration | [45] |
| 2009 i2b2 Medication Challenge | i2b2 | 1,243 de-identified medical discharge summaries annotated for names and details of medications, including dosage, mode, frequency, duration, reason, and presence in a list or narrative structure. | Yes, with registration | [46][47] |
| 2010 i2b2 Relations Challenge | i2b2 | Medical discharge summaries annotated for medical problems, tests, treatments, and the relations among these concepts. Only a subset of these data records are available for research use due to IRB limitations. | Yes, with registration | [6] |
| 2011 i2b2 Coreference Challenge | i2b2 | 978 de-identified medical discharge summaries, progress notes, and other clinical reports annotated with concepts and coreferences. Includes the ODIE corpus. | Yes, with registration | [48] |
| 2012 i2b2 Temporal Relations Challenge | i2b2 | 310 de-identified medical discharge summaries annotated for events and temporal relations. | Yes, with registration | [7] |
| 2014 i2b2 De-identification Challenge | i2b2 | 1,304 de-identified longitudinal medical records annotated for protected health information (PHI). | Yes, with registration | [49] |
| 2014 i2b2 Heart Disease Risk Factors Challenge | i2b2 | 1,304 de-identified longitudinal medical records annotated for risk factors for cardiac artery disease. | Yes, with registration | [50] |
| AIMed | Bunescu et al. | 200 abstracts annotated for protein–protein interactions, as well as negative example abstracts containing no protein-protein interactions. | Yes | [51] |
| BioC-BioGRID | BioCreAtIvE | 120 full text research articles annotated for protein–protein interactions. | Yes | [52] |
| BioCreAtIvE 1 | BioCreAtIvE | 15,000 sentences (10,000 training and 5,000 test) annotated for protein and gene names. 1,000 full text biomedical research articles annotated with protein names and Gene Ontology terms. | Yes | [53] |
| BioCreAtIvE 2 | BioCreAtIvE | 15,000 sentences (10,000 training and 5,000 test, different from the first corpus) annotated for protein and gene names. 542 abstracts linked to EntrezGene identifiers. A variety of research articles annotated for features of protein–protein interactions. | Yes | [54] |
| BioCreative V CDR Task Corpus (BC5CDR) | BioCreAtIvE | 1,500 articles (title and abstract) published in 2014 or later, annotated for 4,409 chemicals, 5,818 diseases and 3116 chemical–disease interactions. | Yes | [55] |
| BioInfer | Pyysalo et al. | 1,100 sentences from biomedical research abstracts annotated for relationships, named entities, and syntactic dependencies. | No | [56] |
| BioScope | Vincze et al. | 1,954 clinical reports, 9 papers, and 1,273 abstracts annotated for linguistic scope and terms denoting negation or uncertainty. | Yes | [57] |
| BioText Recognizing Abbreviation Definitions | BioText Project | 1,000 abstracts on the subject of "yeast", annotated for abbreviations and their meanings. | Yes | [58] |
| BioText Protein–Protein Interaction Data | BioText Project | 1,322 sentences describing protein–protein interactions between HIV-1 and human proteins, annotated with interaction types. | Yes | [59] |
| Comparative Toxicogenomics Database | Davis et al. | A database of manually-curated associations between chemicals, gene products, phenotypes, diseases, and environmental exposures. | Yes | [60] |
| CRAFT | Verspoor et al. | 97 full-text biomedical publications annotated with linguistic structures and biological concepts | Yes | [61] |
| GENIA Corpus | GENIA Project | 1,999 biomedical research abstracts on the topics "human", "blood cells", and "transcription factors", annotated for parts of speech, syntax, terms, events, relations, and coreferences. | Yes | [62][63] |
| FamPlex | Bachman et al. | Protein names and families linked to unique identifiers. Includes affix sets. | Yes | [64] |
| FlySlip Abstracts | FlySlip | 82 research abstracts on Drosophila annotated with gene names. | Yes | [65] |
| FlySlip Full Papers | FlySlip | 5 research papers on Drosophila annotated with anaphoric relations between noun phrases referring to genes and biologically related entities. | Yes | [66] |
| FlySlip Speculative Sentences | FlySlip | More than 1,500 sentences annotated as speculative or not speculative. Includes annotations of clauses. | Yes | [67] |
| IEPA | Ding et al. | 486 sentences from biomedical research abstracts annotated for pairs of co-occurring chemicals, including proteins. | No | [68] |
| JNLPBA corpus | Kim et al. | An extended version of version 3 of the GENIA corpus for NER tasks. | No | [69] |
| Learning Language in Logic (LLL) | Nédellec et al. | 77 sentences from research articles about the bacterium Bacillus subtilis, annotated for protein–gene interactions. | Yes | [70] |
| Medical Subject Headings (MeSH) | National Library of Medicine | Hierarchically-organized terminology for indexing and cataloging biomedical documents. | Yes | [71] |
| Metathesaurus | National Library of Medicine / UMLS | 3.67 million concepts and 14 million concept names, mapped between more than 200 sources of biomedical vocabulary and identifiers. | Yes, with UMLS License Agreement | [72][73] |
| MIMIC-III | MIT Lab for Computational Physiology | de-identified data associated with 53,423 distinct hospital admissions for adult patients. | Requires training and formal access request | [74] |
| ODIE Corpus | Savova et al. | 180 clinical notes annotated with 5,992 coreference pairs. | No | [75] |
| OHSUMED | Hersh et al. | 348,566 biomedical research abstracts and indexing information from MEDLINE, including MeSH (as of 1991). | Yes | [76] |
| PMC Open Access Subset | National Library of Medicine / PubMed Central | More than 2 million research articles, updated weekly. | Yes | [77] |
| RxNorm | National Library of Medicine / UMLS | Normalized names for clinical drugs and drug packs, with combined ingredients, strengths, and form, and assigned types from the Semantic Network. | Yes, with UMLS License Agreement | [78] |
| Semantic Network | National Library of Medicine / UMLS | Lists of 133 semantic types and 54 semantic relationships covering biomedical concepts and vocabulary. | Yes, with UMLS License Agreement | [79][80] |
| SPECIALIST Lexicon | National Library of Medicine / UMLS | A syntactic lexicon of biomedical and general English. | Yes | [81][82] |
| Word Sense Disambiguation (WSD) | National Library of Medicine / UMLS | 203 ambiguous words and 37,888 automatically extracted instances of their use in biomedical research publications. | Yes, with UMLS License Agreement | [83][84] |
| Yapex | Franzén et al. | 200 biomedical research abstracts annotated with protein names. | No | [85] |
Word embeddings
Several groups have developed sets of biomedical vocabulary mapped to vectors of real numbers, known as word vectors or word embeddings. Sources of pre-trained embeddings specific for biomedical vocabulary are listed in the table below. The majority are results of the word2vec model developed by Mikolov et al[86] or variants of word2vec.
| Set Name | Authors or Group | Contents and Source | Citation |
|---|---|---|---|
| BioASQword2vec | BioASQ | Vectors produced by word2vec from 10,876,004 English PubMed abstracts. | [87] |
| bio.nlplab.org resources | Pyysalo et al. | A collection of word vectors produced by different approaches, trained on text from PubMed and PubMed Central. | [88] |
| BioVec | Asgari and Mofrad | Vectors for gene and protein sequences, trained using Swiss-Prot. | [89] |
| RadiologyReportEmbedding | Banerjee et al. | Vectors produced by word2vec from the text of 10,000 radiology reports. | [90] |
Applications
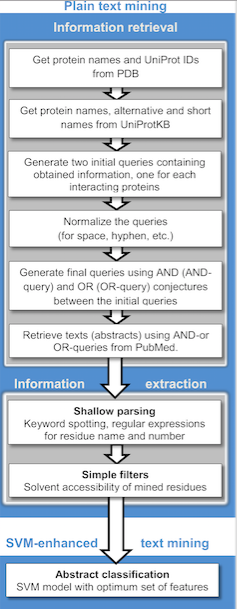
Text mining applications in the biomedical field include computational approaches to assist with studies in protein docking,[91] protein interactions,[92][93] and protein-disease associations.[94] Text mining techniques have several advantages over traditional manual curation for identifying associations. Text mining algorithms can identify and extract information from a vast amount of literature, and more efficiently than manual curation. This includes the integration of data from different sources, including literature, databases, and experimental results. These algorithms have transformed the process of identifying and prioritizing novel genes and gene-disease associations that have previously been overlooked.[95]




These methods are the foundation to facilitate systematic searches of overlooked scientific and biomedical literature which could carry significant association between research. The combination of information can stem new discoveries and hypotheses especially with the integration of datasets. The quality of the database is as important as the size of it. Promising text mining methods such as iProLINK (integrated Protein Literature Information and Knowledge) have been developed to curate data sources that can aid text mining research in areas of bibliography mapping, annotation extraction, protein named entity recognition, and protein ontology development.[96] Curated databases such as UniProt can accelerate the accessibility of targeted information not only for genetic sequences, but also for literature and phylogeny.[citation needed]
Gene cluster identification
Methods for determining the association of gene clusters obtained by microarray experiments with the biological context provided by the corresponding literature have been developed.[97]
Protein interactions
Automatic extraction of protein interactions[98] and associations of proteins to functional concepts (e.g. gene ontology terms) has been explored.[citation needed] The search engine PIE was developed to identify and return protein-protein interaction mentions from MEDLINE-indexed articles.[99] The extraction of kinetic parameters from text or the subcellular location of proteins have also been addressed by information extraction and text mining technology.[citation needed]
Gene-disease associations
Computational gene prioritization is an essential step in understanding the genetic basis of diseases, particularly within genetic linkage analysis. Text mining and other computational tools extract relevant information, including gene-disease associations, among others, from numerous data sources, then apply different ranking algorithms to prioritize the genes based on their relevance to the specific disease.[100] Text mining and gene prioritization allow researchers to focus their efforts on the most promising candidates for further research.
Computational tools for gene prioritization continue to be developed and analyzed. One group studied the performance of various text-mining techniques for disease gene prioritization. They investigated different domain vocabularies, text representation schemes, and ranking algorithms in order to find the best approach for identifying disease-causing genes to establish a benchmark.[101]
Gene-trait associations
An agricultural genomics group identified genes related to bovine reproductive traits using text mining, among other approaches.[102]
Applications of phrase mining to disease associations
A text mining study assembled a collection of 709 core extracellular matrix proteins and associated proteins based on two databases: MatrixDB (matrixdb.univ-lyon1.fr) and UniProt. This set of proteins had a manageable size and a rich body of associated information, making it a suitable for the application of text mining tools. The researchers conducted phrase-mining analysis to cross-examine individual extracellular matrix proteins across the biomedical literature concerned with six categories of cardiovascular diseases. They used a phrase-mining pipeline, Context-aware Semantic Online Analytical Processing (CaseOLAP),[103] then semantically scored all 709 proteins according to their Integrity, Popularity, and Distinctiveness using the CaseOLAP pipeline. The text mining study validated existing relationships and informed previously unrecognized biological processes in cardiovascular pathophysiology.[94]
Software tools
Search engines
Search engines designed to retrieve biomedical literature relevant to a user-provided query frequently rely upon text mining approaches. Publicly available tools specific for research literature include PubMed search, Europe PubMed Central search, GeneView,[104] and APSE[105] Similarly, search engines and indexing systems specific for biomedical data have been developed, including DataMed[106] and OmicsDI.[107]
Some search engines, such as Essie,[108] OncoSearch,[109] PubGene,[110][111] and GoPubMed[112] were previously public but have since been discontinued, rendered obsolete, or integrated into commercial products.
Medical record analysis systems
Electronic medical records (EMRs) and electronic health records (EHRs) are collected by clinical staff in the course of diagnosis and treatment. Though these records generally include structured components with predictable formats and data types, the remainder of the reports are often free-text and difficult to search, leading to challenges with patient care.[113] Numerous complete systems and tools have been developed to analyse these free-text portions.[114] The MedLEE system was originally developed for analysis of chest radiology reports but later extended to other report topics.[115] The clinical Text Analysis and Knowledge Extraction System, or cTAKES, annotates clinical text using a dictionary of concepts.[116] The CLAMP system offers similar functionality with a user-friendly interface.[117]
Frameworks
Computational frameworks have been developed to rapidly build tools for biomedical text mining tasks. SwellShark[118] is a framework for biomedical NER that requires no human-labeled data but does make use of resources for weak supervision (e.g., UMLS semantic types). The SparkText framework[119] uses Apache Spark data streaming, a NoSQL database, and basic machine learning methods to build predictive models from scientific articles.
APIs
Some biomedical text mining and natural language processing tools are available through application programming interfaces, or APIs. NOBLE Coder performs concept recognition through an API.[120]
Conferences
The following academic conferences and workshops host discussions and presentations in biomedical text mining advances. Most publish proceedings.
| Conference Name | Session | Proceedings |
|---|---|---|
| Association for Computational Linguistics (ACL) annual meeting | plenary session and as part of the BioNLP workshop | |
| ACL BioNLP workshop | [121] | |
| American Medical Informatics Association (AMIA) annual meeting | in plenary session | |
| Intelligent Systems for Molecular Biology (ISMB) | in plenary session and in the BioLINK and Bio-ontologies workshops | [122] |
| International Conference on Bioinformatics and Biomedicine (BIBM) | [123] | |
| International Conference on Information and Knowledge Management (CIKM) | within International Workshop on Data and Text Mining in Biomedical Informatics (DTMBIO) | [124] |
| North American Association for Computational Linguistics (NAACL) annual meeting | plenary session and as part of the BioNLP workshop | |
| Pacific Symposium on Biocomputing (PSB) | in plenary session | [125] |
| Practical Applications of Computational Biology & Bioinformatics (PACBB) | [126] | |
| Text REtrieval Conference (TREC) | formerly as part of TREC Genomics track; as of 2018 part of Precision Medicine Track | [127] |
Journals
A variety of academic journals publishing manuscripts on biology and medicine include topics in text mining and natural language processing software. Some journals, including the Journal of the American Medical Informatics Association (JAMIA) and the Journal of Biomedical Informatics are popular publications for these topics.
References
- ^ Jensen, Lars Juhl; Saric, Jasmin; Bork, Peer (February 2006). "Literature mining for the biologist: from information retrieval to biological discovery". Nature Reviews Genetics. 7 (2): 119–129. doi:10.1038/nrg1768. ISSN 1471-0056. PMID 16418747. S2CID 423509.
- ^ Westergaard D, Stærfeldt HH, Tønsberg C, Jensen LJ, Brunak S (February 2018). "A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts". PLOS Computational Biology. 14 (2) e1005962. Bibcode:2018PLSCB..14E5962W. doi:10.1371/journal.pcbi.1005962. PMC 5831415. PMID 29447159.
- ^ Danescu-Niculescu-Mizil C, Lee L (2011). Chameleons in Imagined Conversations: A New Approach to Understanding Coordination of Linguistic Style in Dialogs. pp. 76–87. arXiv:1106.3077. Bibcode:2011arXiv1106.3077D. ISBN 978-1-932432-95-4.
{{cite book}}:|journal=ignored (help) - ^ McAuley J, Leskovec J (2013-10-12). "Hidden factors and hidden topics: Understanding rating dimensions with review text". Proceedings of the 7th ACM conference on Recommender systems. ACM. pp. 165–172. doi:10.1145/2507157.2507163. ISBN 978-1-4503-2409-0. S2CID 6440341.
- ^ a b c Ohno-Machado L, Nadkarni P, Johnson K (2013). "Natural language processing: algorithms and tools to extract computable information from EHRs and from the biomedical literature". Journal of the American Medical Informatics Association. 20 (5): 805. doi:10.1136/amiajnl-2013-002214. PMC 3756279. PMID 23935077.
- ^ a b Uzuner Ö, South BR, Shen S, DuVall SL (2011). "2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text". Journal of the American Medical Informatics Association. 18 (5): 552–6. doi:10.1136/amiajnl-2011-000203. PMC 3168320. PMID 21685143.
- ^ a b Sun W, Rumshisky A, Uzuner O (2013). "Evaluating temporal relations in clinical text: 2012 i2b2 Challenge". Journal of the American Medical Informatics Association. 20 (5): 806–13. doi:10.1136/amiajnl-2013-001628. PMC 3756273. PMID 23564629.
- ^ Stubbs A, Kotfila C, Uzuner Ö (December 2015). "Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1". Journal of Biomedical Informatics. 58 (Suppl): S11–9. doi:10.1016/j.jbi.2015.06.007. PMC 4989908. PMID 26225918.
- ^ Albright D, Lanfranchi A, Fredriksen A, Styler WF, Warner C, Hwang JD, Choi JD, Dligach D, Nielsen RD, Martin J, Ward W, Palmer M, Savova GK (2013). "Towards comprehensive syntactic and semantic annotations of the clinical narrative". Journal of the American Medical Informatics Association. 20 (5): 922–30. doi:10.1136/amiajnl-2012-001317. PMC 3756257. PMID 23355458.
- ^ Bada M, Eckert M, Evans D, Garcia K, Shipley K, Sitnikov D, Baumgartner WA, Cohen KB, Verspoor K, Blake JA, Hunter LE (July 2012). "Concept annotation in the CRAFT corpus". BMC Bioinformatics. 13 (1) 161. doi:10.1186/1471-2105-13-161. PMC 3476437. PMID 22776079.
- ^ Holzinger A, Jurisica I (2014). "Knowledge Discovery and Data Mining in Biomedical Informatics: The Future is in Integrative, Interactive Machine Learning Solutions". Interactive Knowledge Discovery and Data Mining in Biomedical Informatics. Lecture Notes in Computer Science. Vol. 8401. Springer Berlin Heidelberg. pp. 1–18. doi:10.1007/978-3-662-43968-5_1. ISBN 978-3-662-43967-8.
- ^ Ratner A, Bach SH, Ehrenberg H, Fries J, Wu S, Ré C (November 2017). "Snorkel: Rapid Training Data Creation with Weak Supervision". Proceedings of the VLDB Endowment. 11 (3): 269–282. arXiv:1711.10160. Bibcode:2017arXiv171110160R. doi:10.14778/3157794.3157797. PMC 5951191. PMID 29770249.
- ^ Ren X, Wu Z, He W, Qu M, Voss CR, Ji H, Abdelzaher TF, Han J (2017-04-03). "CoType: Joint Extraction of Typed Entities and Relations with Knowledge Bases". Proceedings of the 26th International Conference on World Wide Web. WWW '17. International World Wide Web Conferences Steering Committee. pp. 1015–1024. doi:10.1145/3038912.3052708. ISBN 978-1-4503-4913-0. S2CID 1724837.
- ^ a b Erhardt RA, Schneider R, Blaschke C (April 2006). "Status of text-mining techniques applied to biomedical text". Drug Discovery Today. 11 (7–8): 315–25. doi:10.1016/j.drudis.2006.02.011. PMID 16580973.
- ^ Milosevic N, Gregson C, Hernandez R, Nenadic G (February 2019). "A framework for information extraction from tables in biomedical literature". International Journal on Document Analysis and Recognition. 22 (1): 55–78. arXiv:1902.10031. Bibcode:2019arXiv190210031M. doi:10.1007/s10032-019-00317-0. S2CID 62880746.
- ^ Demner-Fushman D, Shooshan SE, Rodriguez L, Aronson AR, Lang F, Rogers W, Roberts K, Tonning J (January 2018). "A dataset of 200 structured product labels annotated for adverse drug reactions". Scientific Data. 5 180001. Bibcode:2018NatSD...580001D. doi:10.1038/sdata.2018.1. PMC 5789866. PMID 29381145.
- ^ a b Agarwal S, Yu H (December 2010). "Detecting hedge cues and their scope in biomedical text with conditional random fields". Journal of Biomedical Informatics. 43 (6): 953–61. doi:10.1016/j.jbi.2010.08.003. PMC 2991497. PMID 20709188.
- ^ Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V (April 2019). "Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review". JMIR Med Inform. 7 (2) e12239. doi:10.2196/12239. PMC 6528438. PMID 31066697.
- ^ Vandenbussche PY, Cormont S, André C, Daniel C, Delahousse J, Charlet J, Lepage E (2013). "Implementation and management of a biomedical observation dictionary in a large healthcare information system". Journal of the American Medical Informatics Association. 20 (5): 940–6. doi:10.1136/amiajnl-2012-001410. PMC 3756262. PMID 23635601.
- ^ Jannot AS, Zapletal E, Avillach P, Mamzer MF, Burgun A, Degoulet P (June 2017). "The Georges Pompidou University Hospital Clinical Data Warehouse: A 8-years follow-up experience". International Journal of Medical Informatics. 102: 21–28. doi:10.1016/j.ijmedinf.2017.02.006. PMID 28495345.
- ^ Levy B. "Health Care's Semantics Challenge". www.fortherecordmag.com. Great Valley Publishing Company. Retrieved 2018-10-04.
- ^ Goodwin LK, Prather JC (2002). "Protecting patient privacy in clinical data mining". Journal of Healthcare Information Management. 16 (4): 62–7. PMID 12365302.
- ^ Tucker K, Branson J, Dilleen M, Hollis S, Loughlin P, Nixon MJ, Williams Z (July 2016). "Protecting patient privacy when sharing patient-level data from clinical trials". BMC Medical Research Methodology. 16 (S1) 77. doi:10.1186/s12874-016-0169-4. PMC 4943495. PMID 27410040.
- ^ Graves S (2013). "Confidentiality, electronic health records, and the clinician". Perspectives in Biology and Medicine. 56 (1): 105–25. doi:10.1353/pbm.2013.0003. PMID 23748530. S2CID 25816887.
- ^ Leser U, Hakenberg J (2005-01-01). "What makes a gene name? Named entity recognition in the biomedical literature". Briefings in Bioinformatics. 6 (4): 357–369. doi:10.1093/bib/6.4.357. ISSN 1467-5463. PMID 16420734.
- ^ Krallinger M, Leitner F, Rabal O, Vazquez M, Oyarzabal J, Valencia A. "Overview of the chemical compound and drug name recognition (CHEMDNER) task" (PDF). Proceedings of the Fourth BioCreative Challenge Evaluation Workshop. 2: 6–37. Archived from the original on July 14, 2014.
- ^ Jimeno A, Jimenez-Ruiz E, Lee V, Gaudan S, Berlanga R, Rebholz-Schuhmann D (April 2008). "Assessment of disease named entity recognition on a corpus of annotated sentences". BMC Bioinformatics. 9 (Suppl 3) S3. doi:10.1186/1471-2105-9-s3-s3. PMC 2352871. PMID 18426548.
- ^ Habibi M, Weber L, Neves M, Wiegandt DL, Leser U (July 2017). "Deep learning with word embeddings improves biomedical named entity recognition". Bioinformatics. 33 (14): i37–i48. doi:10.1093/bioinformatics/btx228. PMC 5870729. PMID 28881963.
- ^ Furrer L, Cornelius J, Rinaldi F (March 2022). "Parallel sequence tagging for concept recognition". BMC Bioinformatics. 22 (Suppl 1) 623. doi:10.1186/s12859-021-04511-y. PMC 8943923. PMID 35331131.
- ^ Cohen AM (2006). "An effective general purpose approach for automated biomedical document classification". AMIA ... Annual Symposium Proceedings. AMIA Symposium. 2006: 161–5. PMC 1839342. PMID 17238323.
- ^ a b Xu R, Wunsch DC (2010). "Clustering algorithms in biomedical research: a review". IEEE Reviews in Biomedical Engineering. 3: 120–54. Bibcode:2010IRBE....3..120X. doi:10.1109/rbme.2010.2083647. PMID 22275205. S2CID 206522771.
- ^ Rodriguez-Esteban R (December 2009). "Biomedical text mining and its applications". PLOS Computational Biology. 5 (12) e1000597. Bibcode:2009PLSCB...5E0597R. doi:10.1371/journal.pcbi.1000597. PMC 2791166. PMID 20041219.
- ^ Blake C (April 2010). "Beyond genes, proteins, and abstracts: Identifying scientific claims from full-text biomedical articles". Journal of Biomedical Informatics. 43 (2): 173–89. doi:10.1016/j.jbi.2009.11.001. PMID 19900574.
- ^ a b Alamri A, Stevensony M (2015). "Automatic identification of potentially contradictory claims to support systematic reviews". 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE. pp. 930–937. doi:10.1109/bibm.2015.7359808. ISBN 978-1-4673-6799-8. S2CID 28079483.
- ^ Fleuren WW, Alkema W (March 2015). "Application of text mining in the biomedical domain". Methods. 74: 97–106. doi:10.1016/j.ymeth.2015.01.015. PMID 25641519.
- ^ Karp PD (2016-01-01). "Can we replace curation with information extraction software?". Database. 2016 baw150. doi:10.1093/database/baw150. PMC 5199131. PMID 28025341.
- ^ Krallinger M, Valencia A, Hirschman L (2008). "Linking genes to literature: text mining, information extraction, and retrieval applications for biology". Genome Biology. 9 (Suppl 2) S8. doi:10.1186/gb-2008-9-s2-s8. PMC 2559992. PMID 18834499.
- ^ Neves M, Leser U (March 2015). "Question answering for biology". Methods. 74: 36–46. doi:10.1016/j.ymeth.2014.10.023. PMID 25448292.
- ^ Semantics Scholar. (2020) "Cut through the clutter:[Open Access] Download the Coronavirus Open Research Dataset". Semantics Scholar website Retrieved 30 March 2020
- ^ Brennan, Patti. (24 March 2020). "Blog:How Does a Library Respond to a Global Health Crisis?". National Library of Medicine website Retrieved 30 March 2020.
- ^ Brainard J (13 May 2020). "Scientists are drowning in COVID-19 papers. Can new tools keep them afloat?". Science | AAAS. Retrieved 17 May 2020.
- ^ Bossy R, Deléger L, Chaix E, Ba M, Nédellec C (2019). Bacteria biotope at BioNLP open shared tasks 2019. Proceedings of the 5th workshop on BioNLP open shared tasks. Association for Computational Linguistics. pp. 121–131. doi:10.18653/v1/D19-5719.
- ^ Uzuner O, Luo Y, Szolovits P (2007-09-01). "Evaluating the state-of-the-art in automatic de-identification". Journal of the American Medical Informatics Association. 14 (5): 550–63. doi:10.1197/jamia.m2444. PMC 1975792. PMID 17600094.
- ^ Uzuner O, Goldstein I, Luo Y, Kohane I (2008-01-01). "Identifying patient smoking status from medical discharge records". Journal of the American Medical Informatics Association. 15 (1): 14–24. doi:10.1197/jamia.m2408. PMC 2274873. PMID 17947624.
- ^ Uzuner O (2009). "Recognizing obesity and comorbidities in sparse data". Journal of the American Medical Informatics Association. 16 (4): 561–70. doi:10.1197/jamia.M3115. PMC 2705260. PMID 19390096.
- ^ Uzuner O, Solti I, Xia F, Cadag E (2010). "Community annotation experiment for ground truth generation for the i2b2 medication challenge". Journal of the American Medical Informatics Association. 17 (5): 519–23. doi:10.1136/jamia.2010.004200. PMC 2995684. PMID 20819855.
- ^ Uzuner O, Solti I, Cadag E (2010). "Extracting medication information from clinical text". Journal of the American Medical Informatics Association. 17 (5): 514–8. doi:10.1136/jamia.2010.003947. PMC 2995677. PMID 20819854.
- ^ Uzuner O, Bodnari A, Shen S, Forbush T, Pestian J, South BR (2012). "Evaluating the state of the art in coreference resolution for electronic medical records". Journal of the American Medical Informatics Association. 19 (5): 786–91. doi:10.1136/amiajnl-2011-000784. PMC 3422835. PMID 22366294.
- ^ Stubbs A, Uzuner Ö (December 2015). "Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus". Journal of Biomedical Informatics. 58 (Suppl): S20–9. doi:10.1016/j.jbi.2015.07.020. PMC 4978170. PMID 26319540.
- ^ Stubbs A, Uzuner Ö (December 2015). "Annotating risk factors for heart disease in clinical narratives for diabetic patients". Journal of Biomedical Informatics. 58 (Suppl): S78–91. doi:10.1016/j.jbi.2015.05.009. PMC 4978180. PMID 26004790.
- ^ Bunescu R, Ge R, Kate RJ, Marcotte EM, Mooney RJ, Ramani AK, Wong YW (February 2005). "Comparative experiments on learning information extractors for proteins and their interactions". Artificial Intelligence in Medicine. 33 (2): 139–55. CiteSeerX 10.1.1.10.2168. doi:10.1016/j.artmed.2004.07.016. PMID 15811782.
- ^ Islamaj Dogan R, Kim S, Chatr-Aryamontri A, Chang CS, Oughtred R, Rust J, Wilbur WJ, Comeau DC, Dolinski K, Tyers M (2017-01-01). "The BioC-BioGRID corpus: full text articles annotated for curation of protein-protein and genetic interactions". Database. 2017 baw147. doi:10.1093/database/baw147. PMC 5225395. PMID 28077563.
- ^ Hirschman L, Yeh A, Blaschke C, Valencia A (2005). "Overview of BioCreAtIvE: critical assessment of information extraction for biology". BMC Bioinformatics. 6 (Suppl 1) S1. doi:10.1186/1471-2105-6-S1-S1. PMC 1869002. PMID 15960821.
- ^ Krallinger M, Morgan A, Smith L, Leitner F, Tanabe L, Wilbur J, Hirschman L, Valencia A (2008). "Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge". Genome Biology. 9 (Suppl 2) S1. doi:10.1186/gb-2008-9-s2-s1. PMC 2559980. PMID 18834487.
- ^ Li J, Sun Y, Johnson RJ, Sciaky D, Wei CH, Leaman R, Davis AP, Mattingly CJ, Wiegers TC, Lu Z (2016). "BioCreative V CDR task corpus: a resource for chemical disease relation extraction". Database. 2016 baw068. doi:10.1093/database/baw068. PMC 4860626. PMID 27161011.
- ^ Pyysalo S, Ginter F, Heimonen J, Björne J, Boberg J, Järvinen J, Salakoski T (February 2007). "BioInfer: a corpus for information extraction in the biomedical domain". BMC Bioinformatics. 8 (1) 50. doi:10.1186/1471-2105-8-50. PMC 1808065. PMID 17291334.
- ^ Vincze V, Szarvas G, Farkas R, Móra G, Csirik J (November 2008). "The BioScope corpus: biomedical texts annotated for uncertainty, negation and their scopes". BMC Bioinformatics. 9 (Suppl 11) S9. doi:10.1186/1471-2105-9-s11-s9. PMC 2586758. PMID 19025695.
- ^ Schwartz AS, Hearst MA (2003). "A simple algorithm for identifying abbreviation definitions in biomedical text". Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing: 451–62. PMID 12603049.
- ^ Rosario B, Hearst MA (2005-10-06). "Multi-way relation classification". Multi-way relation classification: application to protein-protein interactions. Hlt '05. Association for Computational Linguistics. pp. 732–739. doi:10.3115/1220575.1220667. S2CID 902226.
- ^ Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, et al. (January 2019). "The Comparative Toxicogenomics Database: update 2019". Nucleic Acids Research. 47 (D1): D948–D954. doi:10.1093/nar/gky868. PMC 6323936. PMID 30247620.
- ^ Verspoor K, Cohen KB, Lanfranchi A, Warner C, Johnson HL, Roeder C, Choi JD, Funk C, Malenkiy Y, Eckert M, Xue N, Baumgartner WA, Bada M, Palmer M, Hunter LE (August 2012). "A corpus of full-text journal articles is a robust evaluation tool for revealing differences in performance of biomedical natural language processing tools". BMC Bioinformatics. 13 (1) 207. doi:10.1186/1471-2105-13-207. PMC 3483229. PMID 22901054.
- ^ Kim JD, Ohta T, Tateisi Y, Tsujii J (2003-07-03). "GENIA corpus--a semantically annotated corpus for bio-textmining". Bioinformatics. 19 (Suppl 1): i180–i182. doi:10.1093/bioinformatics/btg1023. PMID 12855455.
- ^ "GENIA Project". www.geniaproject.org. Retrieved 2018-10-06.
- ^ Bachman JA, Gyori BM, Sorger PK (June 2018). "FamPlex: a resource for entity recognition and relationship resolution of human protein families and complexes in biomedical text mining". BMC Bioinformatics. 19 (1) 248. doi:10.1186/s12859-018-2211-5. PMC 6022344. PMID 29954318.
- ^ Vlachos A, Gasperin C (2006). "Bootstrapping and evaluating named entity recognition in the biomedical domain". BioNLP '06 Proceedings of the Workshop on Linking Natural Language Processing and Biology: Towards Deeper Biological Literature Analysis. BioNLP '06: 138–145. doi:10.3115/1567619.1567652.
- ^ Gasperin C, Karamanis N, Seal R (2007). "Annotation of anaphoric relations in biomedical full text articles using a domain-relevant scheme". Proceedings of DAARC 2007: 19–24.
- ^ Medlock B, Briscoe T (2007). "Weakly Supervised Learning for Hedge Classification in Scientific Literature" (PDF). Proceedings of the 45th Annual Meeting of the Association of Computational Linguistics: 992–999.
- ^ Ding J, Berleant D, Nettleton D, Wurtele E (2001). "Mining MEDLINE: Abstracts, sentences, or phrases?". In Altman RB, Dunker AK, Hunter L, Lauderdale K, Klein TE (eds.). Pacific Symposium on Biocomputing 2002. World Scientific. pp. 326–337. CiteSeerX 10.1.1.385.6071. doi:10.1142/9789812799623_0031. ISBN 978-981-02-4777-5. PMID 11928487.
{{cite book}}:|journal=ignored (help) - ^ Kim J, Ohta T, Tsuruoka Y, Tateisi Y, Collier N (2004). "Introduction to the bio-entity recognition task at JNLPBA". Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications - JNLPBA '04: 70. doi:10.3115/1567594.1567610.
- ^ "LLLchallenge". genome.jouy.inra.fr. Retrieved 2018-10-06.
- ^ "Medical Subject Headings - Home Page". www.nlm.nih.gov. Retrieved 2018-10-06.
- ^ Bodenreider O (January 2004). "The Unified Medical Language System (UMLS): integrating biomedical terminology". Nucleic Acids Research. 32 (Database issue): D267–70. doi:10.1093/nar/gkh061. PMC 308795. PMID 14681409.
- ^ "Metathesaurus". www.nlm.nih.gov. Retrieved 2018-10-07.
- ^ Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG (May 2016). "MIMIC-III, a freely accessible critical care database". Scientific Data. 3 160035. Bibcode:2016NatSD...360035J. doi:10.1038/sdata.2016.35. PMC 4878278. PMID 27219127.
- ^ Savova GK, Chapman WW, Zheng J, Crowley RS (2011). "Anaphoric relations in the clinical narrative: corpus creation". Journal of the American Medical Informatics Association. 18 (4): 459–65. doi:10.1136/amiajnl-2011-000108. PMC 3128403. PMID 21459927.
- ^ Hersh W, Buckley C, Leone TJ, Hickam D (1994). "OHSUMED: An Interactive Retrieval Evaluation and New Large Test Collection for Research". Sigir '94. Springer London. pp. 192–201. doi:10.1007/978-1-4471-2099-5_20. ISBN 978-3-540-19889-5. S2CID 15094383.
- ^ "Open Access Subset". www.ncbi.nlm.nih.gov. Retrieved 2018-10-06.
- ^ Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R (2011). "Normalized names for clinical drugs: RxNorm at 6 years". Journal of the American Medical Informatics Association. 18 (4): 441–8. doi:10.1136/amiajnl-2011-000116. PMC 3128404. PMID 21515544.
- ^ McCray AT (2003). "An upper-level ontology for the biomedical domain". Comparative and Functional Genomics. 4 (1): 80–4. doi:10.1002/cfg.255. PMC 2447396. PMID 18629109.
- ^ "The UMLS Semantic Network". semanticnetwork.nlm.nih.gov. Retrieved 2018-10-07.
- ^ McCray AT, Srinivasan S, Browne AC (1994). "Lexical methods for managing variation in biomedical terminologies". Proceedings. Symposium on Computer Applications in Medical Care: 235–9. PMC 2247735. PMID 7949926.
- ^ "The SPECIALIST NLP Tools". lexsrv3.nlm.nih.gov. Retrieved 2018-10-07.
- ^ Jimeno-Yepes AJ, McInnes BT, Aronson AR (June 2011). "Exploiting MeSH indexing in MEDLINE to generate a data set for word sense disambiguation". BMC Bioinformatics. 12 (1) 223. doi:10.1186/1471-2105-12-223. PMC 3123611. PMID 21635749.
- ^ "Word Sense Disambiguation (WSD) Test Collections". wsd.nlm.nih.gov. Archived from the original on 2024-12-13. Retrieved 2018-10-07.
- ^ Franzén K, Eriksson G, Olsson F, Asker L, Lidén P, Cöster J (December 2002). "Protein names and how to find them". International Journal of Medical Informatics. 67 (1–3): 49–61. CiteSeerX 10.1.1.14.2183. doi:10.1016/s1386-5056(02)00052-7. PMID 12460631.
- ^ Mikolov T, Chen K, Corrado G, Dean J (2013-01-16). "Efficient Estimation of Word Representations in Vector Space". arXiv:1301.3781 [cs.CL].
- ^ "BioASQ Releases Continuous Space Word Vectors Obtained by Applying Word2Vec to PubMed Abstracts | bioasq.org". bioasq.org. Retrieved 2018-11-07.
- ^ "bio.nlplab.org". bio.nlplab.org. Retrieved 2018-11-07.
- ^ Asgari E, Mofrad MR (2015-11-10). "Continuous Distributed Representation of Biological Sequences for Deep Proteomics and Genomics". PLOS ONE. 10 (11) e0141287. arXiv:1503.05140. Bibcode:2015PLoSO..1041287A. doi:10.1371/journal.pone.0141287. PMC 4640716. PMID 26555596.
- ^ Banerjee I, Madhavan S, Goldman RE, Rubin DL (2017). "Intelligent Word Embeddings of Free-Text Radiology Reports". AMIA ... Annual Symposium Proceedings. AMIA Symposium. 2017: 411–420. arXiv:1711.06968. Bibcode:2017arXiv171106968B. PMC 5977573. PMID 29854105.
- ^ a b Badal VD, Kundrotas PJ, Vakser IA (December 2015). "Text Mining for Protein Docking". PLOS Computational Biology. 11 (12) e1004630. Bibcode:2015PLSCB..11E4630B. doi:10.1371/journal.pcbi.1004630. PMC 4674139. PMID 26650466.
- ^ Papanikolaou N, Pavlopoulos GA, Theodosiou T, Iliopoulos I (March 2015). "Protein-protein interaction predictions using text mining methods". Methods. 74: 47–53. doi:10.1016/j.ymeth.2014.10.026. PMID 25448298.
- ^ Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (January 2017). "The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible". Nucleic Acids Research. 45 (D1): D362–D368. doi:10.1093/nar/gkw937. PMC 5210637. PMID 27924014.
- ^ a b Liem DA, Murali S, Sigdel D, Shi Y, Wang X, Shen J, Choi H, Caufield JH, Wang W, Ping P, Han J (October 2018). "Phrase mining of textual data to analyze extracellular matrix protein patterns across cardiovascular disease". American Journal of Physiology. Heart and Circulatory Physiology. 315 (4): H910–H924. doi:10.1152/ajpheart.00175.2018. PMC 6230912. PMID 29775406.
- ^ Yu S, Tranchevent LC, De Moor B, Moreau Y (January 2010). "Gene prioritization and clustering by multi-view text mining". BMC Bioinformatics. 11 (1) 28. doi:10.1186/1471-2105-11-28. PMC 3098068. PMID 20074336.
- ^ Hu, Zhang-Zhi; Mani, Inderjeet; Hermoso, Vincent; Liu, Hongfang; Wu, Cathy H. (December 2004). "iProLINK: an integrated protein resource for literature mining". Computational Biology and Chemistry. 28 (5–6): 409–416. doi:10.1016/j.compbiolchem.2004.09.010. PMID 15556482.
- ^ Kankar P, Adak S, Sarkar A, Murari K, Sharma G (11 April 2002). MedMeSH summarizer: text mining for gene clusters. InProceedings of the 2002 SIAM International Conference on Data Mining. Society for Industrial and Applied Mathematics. pp. 548–565. CiteSeerX 10.1.1.215.6230. doi:10.1137/1.9781611972726.32. ISBN 978-0-89871-517-0.
- ^ Pyysalo S, Airola A, Heimonen J, Björne J, Ginter F, Salakoski T (April 2008). "Comparative analysis of five protein-protein interaction corpora". BMC Bioinformatics. 9 (Suppl 3) S6. doi:10.1186/1471-2105-9-s3-s6. PMC 2349296. PMID 18426551.
- ^ Kim S, Kwon D, Shin SY, Wilbur WJ (February 2012). "PIE the search: searching PubMed literature for protein interaction information". Bioinformatics. 28 (4): 597–8. doi:10.1093/bioinformatics/btr702. PMC 3278758. PMID 22199390.
- ^ Gill N, Singh S, Aseri TC (June 2014). "Computational disease gene prioritization: an appraisal". Journal of Computational Biology. 21 (6): 456–465. doi:10.1089/cmb.2013.0158. PMID 24665902.
- ^ Yu S, Van Vooren S, Tranchevent LC, De Moor B, Moreau Y (August 2008). "Comparison of vocabularies, representations and ranking algorithms for gene prioritization by text mining". Bioinformatics. 24 (16): i119–25. doi:10.1093/bioinformatics/btn291. PMID 18689812.
- ^ Hulsegge I, Woelders H, Smits M, Schokker D, Jiang L, Sørensen P (May 2013). "Prioritization of candidate genes for cattle reproductive traits, based on protein-protein interactions, gene expression, and text-mining". Physiological Genomics. 45 (10): 400–6. doi:10.1152/physiolgenomics.00172.2012. PMID 23572538.
- ^ Tao F, Zhuang H, Yu CW, Wang Q, Cassidy T, Kaplan LR, Voss CR, Han J (2016). "Multi-Dimensional, Phrase-Based Summarization in Text Cubes" (PDF). IEEE Data Eng. Bull. 39 (3): 74–84.
- ^ Thomas P, Starlinger J, Vowinkel A, Arzt S, Leser U (July 2012). "GeneView: a comprehensive semantic search engine for PubMed". Nucleic Acids Research. 40 (Web Server issue): W585–91. doi:10.1093/nar/gks563. PMC 3394277. PMID 22693219.
- ^ Brown P, Zhou Y (September 2017). "Biomedical literature: Testers wanted for article search tool". Nature. 549 (7670): 31. Bibcode:2017Natur.549...31B. doi:10.1038/549031c. hdl:10072/408393. PMID 28880292.
- ^ Ohno-Machado L, Sansone SA, Alter G, Fore I, Grethe J, Xu H, Gonzalez-Beltran A, Rocca-Serra P, Gururaj AE, Bell E, Soysal E, Zong N, Kim HE (May 2017). "Finding useful data across multiple biomedical data repositories using DataMed". Nature Genetics. 49 (6): 816–819. doi:10.1038/ng.3864. PMC 6460922. PMID 28546571.
- ^ Perez-Riverol Y, Bai M, da Veiga Leprevost F, Squizzato S, Park YM, Haug K, et al. (May 2017). "Discovering and linking public omics data sets using the Omics Discovery Index". Nature Biotechnology. 35 (5): 406–409. doi:10.1038/nbt.3790. PMC 5831141. PMID 28486464.
- ^ Ide NC, Loane RF, Demner-Fushman D (2007-05-01). "Essie: a concept-based search engine for structured biomedical text". Journal of the American Medical Informatics Association. 14 (3): 253–63. doi:10.1197/jamia.m2233. PMC 2244877. PMID 17329729.
- ^ Lee HJ, Dang TC, Lee H, Park JC (July 2014). "OncoSearch: cancer gene search engine with literature evidence". Nucleic Acids Research. 42 (Web Server issue): W416–21. doi:10.1093/nar/gku368. PMC 4086113. PMID 24813447.
- ^ Jenssen TK, Laegreid A, Komorowski J, Hovig E (May 2001). "A literature network of human genes for high-throughput analysis of gene expression". Nature Genetics. 28 (1): 21–8. doi:10.1038/ng0501-21. PMID 11326270. S2CID 8889284.
- ^ Masys DR (May 2001). "Linking microarray data to the literature". Nature Genetics. 28 (1): 9–10. doi:10.1038/ng0501-9. PMID 11326264. S2CID 52848745.
- ^ Doms A, Schroeder M (July 2005). "GoPubMed: exploring PubMed with the Gene Ontology". Nucleic Acids Research. 33 (Web Server issue): W783–6. doi:10.1093/nar/gki470. PMC 1160231. PMID 15980585.
- ^ Turchin A, Florez Builes LF (May 2021). "Using Natural Language Processing to Measure and Improve Quality of Diabetes Care: A Systematic Review". Journal of Diabetes Science and Technology. 15 (3): 553–560. doi:10.1177/19322968211000831. PMC 8120048. PMID 33736486.
- ^ Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, et al. (January 2018). "Clinical information extraction applications: A literature review". Journal of Biomedical Informatics. 77: 34–49. doi:10.1016/j.jbi.2017.11.011. PMC 5771858. PMID 29162496.
- ^ Friedman C (1997). "Towards a comprehensive medical language processing system: methods and issues". Proceedings: 595–599. PMC 2233560. PMID 9357695.
- ^ Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG (2010). "Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications". Journal of the American Medical Informatics Association. 17 (5): 507–513. doi:10.1136/jamia.2009.001560. PMC 2995668. PMID 20819853.
- ^ Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, Xu H (March 2018). "CLAMP - a toolkit for efficiently building customized clinical natural language processing pipelines". Journal of the American Medical Informatics Association. 25 (3): 331–336. doi:10.1093/jamia/ocx132. PMC 7378877. PMID 29186491.
- ^ Fries J, Wu S, Ratner A, Ré C (2017-04-20). "SwellShark: A Generative Model for Biomedical Named Entity Recognition without Labeled Data". arXiv:1704.06360 [cs.CL].
- ^ Ye Z, Tafti AP, He KY, Wang K, He MM (2016-09-29). "SparkText: Biomedical Text Mining on Big Data Framework". PLOS ONE. 11 (9) e0162721. Bibcode:2016PLoSO..1162721Y. doi:10.1371/journal.pone.0162721. PMC 5042555. PMID 27685652.
- ^ Tseytlin E, Mitchell K, Legowski E, Corrigan J, Chavan G, Jacobson RS (January 2016). "NOBLE - Flexible concept recognition for large-scale biomedical natural language processing". BMC Bioinformatics. 17 (1) 32. doi:10.1186/s12859-015-0871-y. PMC 4712516. PMID 26763894.
- ^ "BioNLP - ACL Anthology". aclanthology.coli.uni-saarland.de. Archived from the original on 2018-10-18. Retrieved 2018-10-17.
- ^ "ISMB Proceedings". www.iscb.org. Retrieved 2018-10-18.
- ^ "All Proceedings". ieeexplore.ieee.org. Retrieved 2025-01-19.
- ^ "dblp: CIKM". dblp.uni-trier.de. Retrieved 2018-10-17.
- ^ "PSB Proceedings". psb.stanford.edu. Retrieved 2018-10-18.
- ^ "dblp: Practical Applications of Computational Biology & Bioinformatics". dblp.org. Retrieved 2018-10-17.
- ^ "Text REtrieval Conference (TREC) Proceedings". trec.nist.gov. Retrieved 2018-10-17.
Further reading
- Krallinger M, Valencia A (2005). "Text-mining and information-retrieval services for molecular biology". Genome Biology. 6 (7): 224. doi:10.1186/gb-2005-6-7-224. PMC 1175978. PMID 15998455.
- Hoffmann R, Krallinger M, Andres E, Tamames J, Blaschke C, Valencia A (May 2005). "Text mining for metabolic pathways, signaling cascades, and protein networks". Science's STKE. 2005 (283) pe21. doi:10.1126/stke.2832005pe21. PMID 15886388. S2CID 15301069.
- Krallinger M, Erhardt RA, Valencia A (March 2005). "Text-mining approaches in molecular biology and biomedicine". Drug Discovery Today. 10 (6): 439–45. doi:10.1016/S1359-6446(05)03376-3. PMID 15808823.
- Biomedical Literature Mining Publications (BLIMP) Archived 2004-08-29 at the Wayback Machine: A comprehensive and regularly updated index of publications on (bio)medical text mining
External links
- Bio-NLP resources, systems and application database collection Archived 2009-05-04 at the Wayback Machine
- The BioNLP mailing list archives
- Corpora for biomedical text mining Archived 2011-07-24 at the Wayback Machine
- The BioCreative evaluations of biomedical text mining technologies
- Directory of people involved in BioNLP Archived 2011-08-09 at the Wayback Machine
Notes
This article is a direct transclusion of the Wikipedia article and therefore may not meet the same editing standards as LIMSwiki.









