Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text) |
Shawndouglas (talk | contribs) (Updated article of the week text) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1.1 Davis FrontBioinfo2022 40.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:ApE, A Plasmid Editor: A freely available DNA manipulation and visualization program|ApE, A Plasmid Editor: A freely available DNA manipulation and visualization program]]"''' | ||
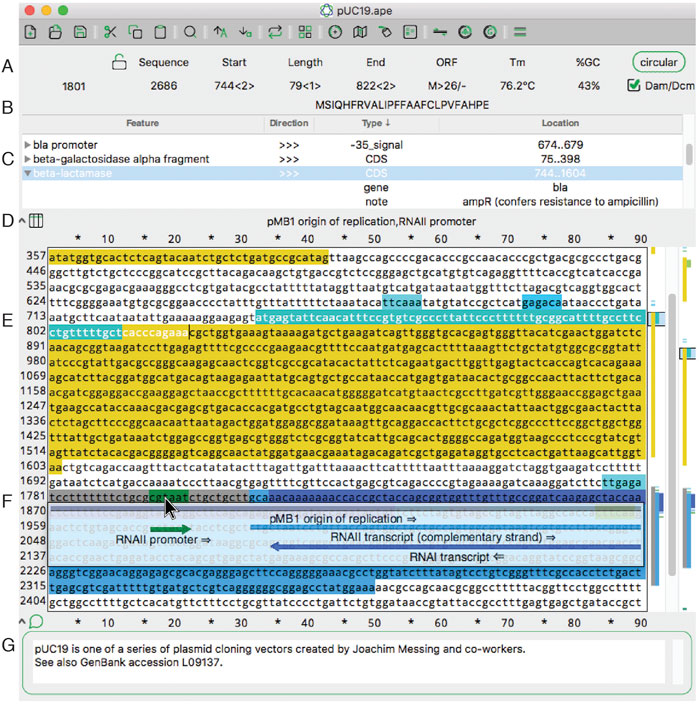
A Plasmid Editor (ApE) is a free, multi-platform application for [[Data visualization|visualizing]], designing, and presenting biologically relevant [[DNA sequencing|DNA sequences]]. ApE provides a flexible framework for annotating a sequence manually or using a user-defined library of features. ApE can be used in designing [[plasmid]]s and other constructs via ''in silico'' simulation of cloning methods such as [[polymerase chain reaction]] (PCR), Gibson assembly, restriction-ligation assembly, and Golden Gate assembly. In addition, ApE provides a platform for creating visually appealing linear and circular plasmid maps. It is available for Mac, PC, and Linux-based platforms and can be downloaded at https://jorgensen.biology.utah.edu/wayned/ape/. ('''[[Journal:ApE, A Plasmid Editor: A freely available DNA manipulation and visualization program|Full article...]]''')<br /> | |||
''Recently featured'': | ''Recently featured'': | ||
{{flowlist | | {{flowlist | | ||
* [[Journal:Development and national scale implementation of an open-source electronic laboratory information system (OpenELIS) in Côte d’Ivoire: Sustainability lessons from the first 13 years|Development and national scale implementation of an open-source electronic laboratory information system (OpenELIS) in Côte d’Ivoire: Sustainability lessons from the first 13 years]] | |||
* [[Journal:AI4Green: An open-source ELN for green and sustainable chemistry|AI4Green: An open-source ELN for green and sustainable chemistry]] | * [[Journal:AI4Green: An open-source ELN for green and sustainable chemistry|AI4Green: An open-source ELN for green and sustainable chemistry]] | ||
* [[Journal:The modeling of laboratory information systems in higher education based on enterprise architecture planning (EAP) for optimizing monitoring and equipment maintenance|The modeling of laboratory information systems in higher education based on enterprise architecture planning (EAP) for optimizing monitoring and equipment maintenance]] | * [[Journal:The modeling of laboratory information systems in higher education based on enterprise architecture planning (EAP) for optimizing monitoring and equipment maintenance|The modeling of laboratory information systems in higher education based on enterprise architecture planning (EAP) for optimizing monitoring and equipment maintenance]] | ||
}} | }} | ||
Revision as of 16:13, 2 October 2023
"ApE, A Plasmid Editor: A freely available DNA manipulation and visualization program"
A Plasmid Editor (ApE) is a free, multi-platform application for visualizing, designing, and presenting biologically relevant DNA sequences. ApE provides a flexible framework for annotating a sequence manually or using a user-defined library of features. ApE can be used in designing plasmids and other constructs via in silico simulation of cloning methods such as polymerase chain reaction (PCR), Gibson assembly, restriction-ligation assembly, and Golden Gate assembly. In addition, ApE provides a platform for creating visually appealing linear and circular plasmid maps. It is available for Mac, PC, and Linux-based platforms and can be downloaded at https://jorgensen.biology.utah.edu/wayned/ape/. (Full article...)
Recently featured:
- Development and national scale implementation of an open-source electronic laboratory information system (OpenELIS) in Côte d’Ivoire: Sustainability lessons from the first 13 years
- AI4Green: An open-source ELN for green and sustainable chemistry
- The modeling of laboratory information systems in higher education based on enterprise architecture planning (EAP) for optimizing monitoring and equipment maintenance