Journal:Ten simple rules to enable multi-site collaborations through data sharing
| Full article title | Ten simple rules to enable multi-site collaborations through data sharing |
|---|---|
| Journal | PLOS Computational Biology |
| Author(s) | Boland, Mary Regina; Karczewski, Konrad J.; Tatonetti, Nicholas P. |
| Author affiliation(s) | Columbia University (NY), Broad Institute of MIT and Harvard, Massachusetts General Hospital |
| Primary contact | Email: mary dot boland @ columbia dot edu |
| Year published | 2017 |
| Volume and issue | 13(1) |
| Page(s) | e1005278 |
| DOI | 10.1371/journal.pcbi.1005278 |
| ISSN | 1553-7358 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005278 |
| Download | http://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1005278&type=printable (PDF) |
Introduction
Open access, open data, and software are critical for advancing science and enabling collaboration across multiple institutions and throughout the world. Despite near universal recognition of its importance, major barriers still exist to sharing raw data, software, and research products throughout the scientific community. Many of these barriers vary by specialty[1], increasing the difficulties for interdisciplinary and/or translational researchers to engage in collaborative research. Multi-site collaborations are vital for increasing both the impact and the generalizability of research results. However, they often present unique data sharing challenges. We discuss enabling multi-site collaborations through enhanced data sharing in this set of Ten Simple Rules.
Collaboration is an essential component of research[2] that takes many forms, including internal (across departments within a single institution) and external collaborations (across institutions). However, multi-site collaborations with more than two institutions encounter more complex challenges because of institutional-specific restrictions and guidelines.[3] Vicens and Bourne focus on collaborators working together on a shared research grant.[4] They do not discuss the specific complexities of multi-site collaborations and the vital need for enhanced data sharing in the multi-site and large-scale collaboration context, in which participants may or may not have the same funding source and/or research grant.
While challenging, multi-site collaborations are equally rewarding and result in increased research productivity.[5][6] One highly successful multi-site and translational collaboration is the Electronic Medical Records and Genomics (eMERGE) network (URL: https://emerge.mc.vanderbilt.edu/) initiated in 2007.[7] The eMERGE network links biorepository data with clinical information from electronic health records (EHRs). They were able to find novel associations and replicate many known associations between genetic variants and clinical phenotypes that would have been more difficult without the collaboration.[8] eMERGE members also collaborated with other consortiums and networks, including the Alzheimer’s Disease Genetics Consortium[9] and the NINDS Stroke Genetics Network[10], to name a few. Other successful collaborations include OHDSI: Observational Health Data Sciences and Informatics (http://www.ohdsi.org/), which builds off of the methodology from the Observational Medical Outcomes Partnership (OMOP)[11], and CIRCLE: Clinical Informatics Research Collaborative (http://circleinformatics.org/). In genetics, there are many consortiums, including ExAC: The Exome Aggregation Consortium (http://exac.broadinstitute.org/), the 1000 Genomes Project Consortium (http://www.1000genomes.org/), the Australian BioGRID (https://www.biogrid.org.au/), The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/), Genotype-Tissue Expression Portal (GTEx: http://www.gtexportal.org/home/), and Encyclopedia of DNA Elements at UCSC (ENCODE: https://genome.ucsc.edu/ENCODE/) among others.
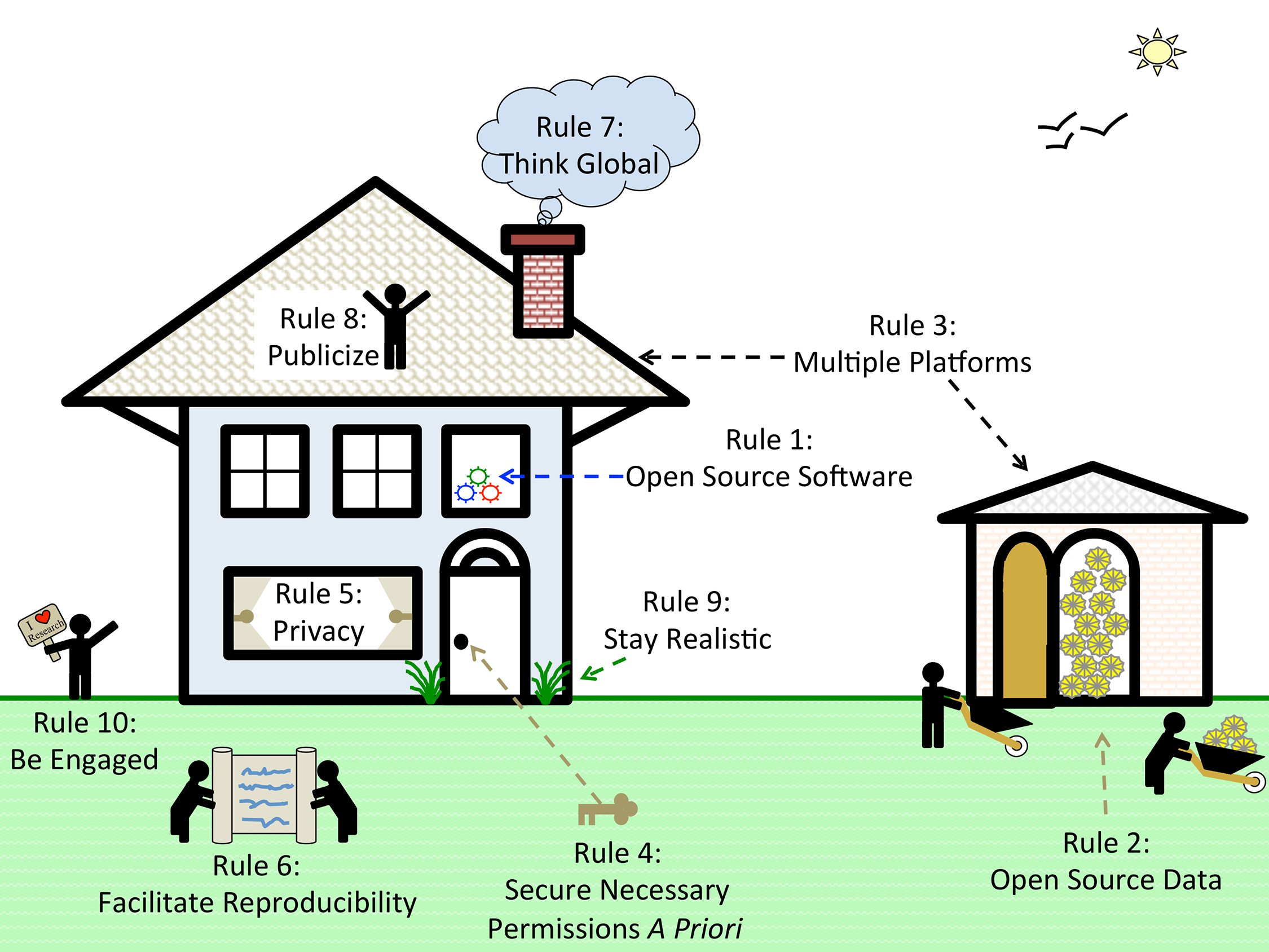
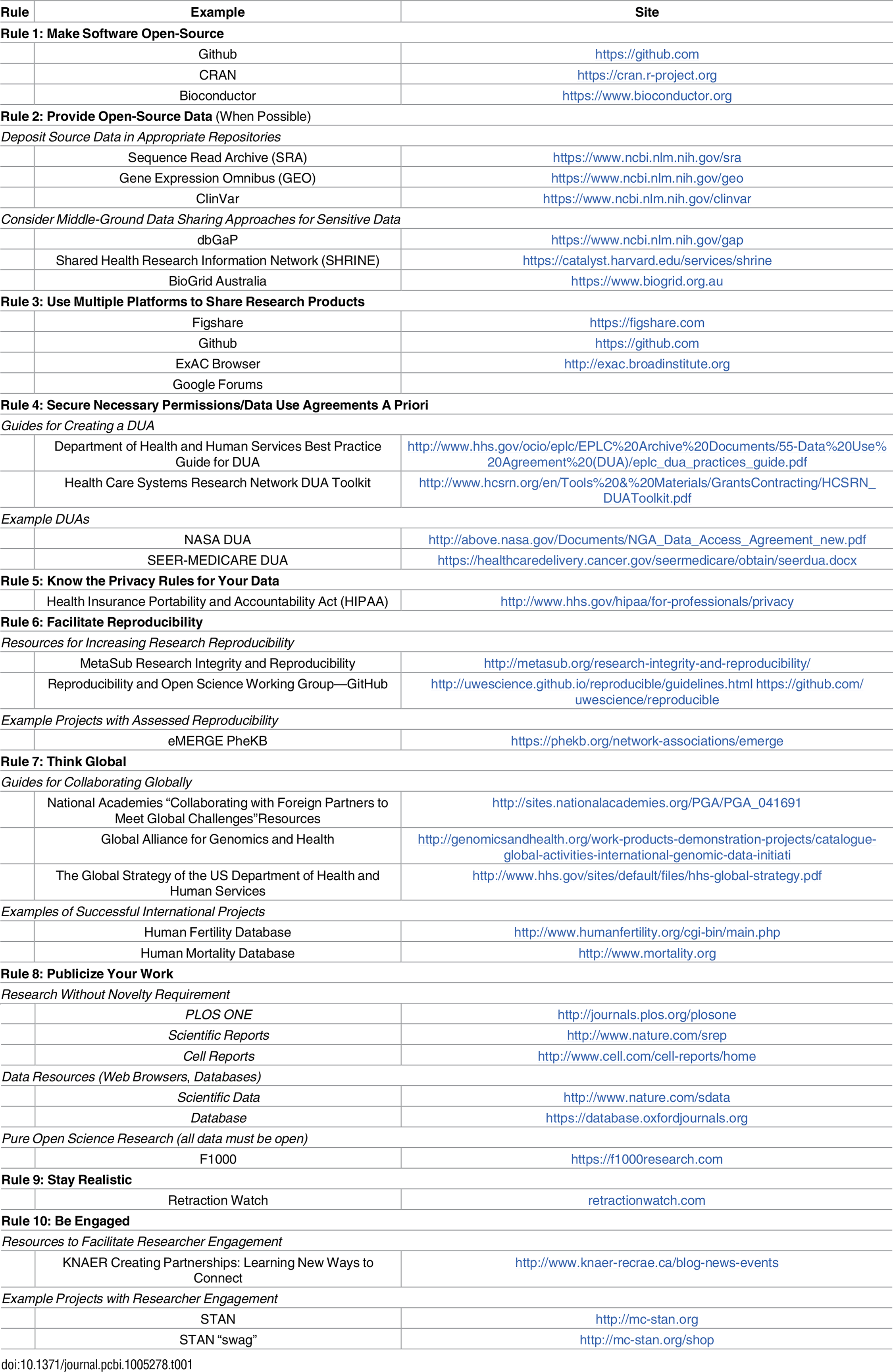
Based on our experiences as both users and participants in collaborations, we present 10 simple rules on how to enable multi-site collaborations within the scientific community through enhanced data sharing. The rules focus on understanding privacy constraints, utilizing proper platforms to facilitate data sharing, thinking in global terms, and encouraging researcher engagement through incentives. We present these 10 rules in the form of a pictograph of modern life (Fig. 1), and we provide a table of example sources and sites that can be referred to for each of the ten rules (Table 1). Please note that this table is not meant to be exhaustive, only to provide some sample resources of use to the research community.
|
|
Definitions
In this paper, we use the term "research product" to include all results from research. This includes algorithms, developed software tools, databases, raw source data, cleaned data, and various metadata generated as a result of the research activity. We differentiate this from "data," which comprises the primary "facts and statistics collected together for analysis" for that particular collaboration. Therefore, data could include genetic data or clinical data. By these definitions, developed software tools are not "data" but "research products." Novel genetic sequences collected for analysis would be considered "raw source data," which is a type of "research product."
Rule 1: Make software open-source
The cornerstone of facilitating multi-site collaborations is to enhance data sharing and make software open-source.[12] By allowing the source code to be open, researchers allow others to both reproduce their work and build upon it in novel ways. To engage in multi-site collaborations, it is necessary for collaborators to have access to code in a repository that is shared among collaborators (although, this could be private and not open to the general public). When the study is complete and the paper is under review and/or published, a stable copy of the code should be made available to the general public. Internal sharing allows the code to be developed, while public sharing of a stable version allows the code to be refined and built upon by others.
Many researchers still limit access to their work despite the known advantages of making software open-source upon publication (e.g., higher impact publications[5]). For example, they allow users to interact with their algorithm by inputting data and receiving results on a web platform, while the backend algorithm often remains inaccessible. Masum et al. advocate the reuse of existing code in their Ten Simple Rules for cultivating open science.[13] However, this is often easier said than done. As long as the back-end algorithms remain hidden, open science will not be possible. Therefore, it is essential for researchers interested in participating in multi-site collaborations to make their software code and algorithms open. Because making software truly "open" can be complex, Prlić and Proctor provide Ten Simple Rules to assist researchers in making their software open-source.[12] Truly open-source software is an essential component in collaborations.[13] Openness also has advantages for the researchers themselves. With more eyes on the source code, others within the community can refine the code, leading to greater identification and correction of errors. There are several methods for sharing software code. If you use the R platform, then libraries can be shared with the entire open-source community via CRAN (https://cran.r-project.org/) and Bioconductor, which is specifically for biologically related algorithms (https://www.bioconductor.org/). Code can also be shared on Github with issue trackers for error detection.
Rule 2: Provide open-source data
Deposit source data in appropriate repositories
Whenever possible, it is important to make source data available. Openness benefits your collaborators by allowing them to perform additional analyses easily. Source data could include not only processed or cleaned data used in algorithms but also raw data files. These files can often be very large; therefore, they are often stored in some external site or data warehouse. The National Center for Biotechnology Information (NCBI) maintains the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) and the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/); both are great places to deposit source data, if appropriate.
In addition to raw data files, it is also helpful to provide intermediate data files at various stages of processing. If comparing your results to those in the literature, it can also be useful to provide a meta-analysis with publications (along with PubMed IDs) that detail those publications that support and refute the results you obtained.
Data sharing is vitally important for multi-site collaborations by allowing researchers to compare results from across vastly different study populations, which increases the generalizability of the findings.[14] While a multi-site research project is still ongoing, data can be shared in a private shared space until all necessary data quality checks have been conducted and the findings have been published. After publication, data can be deposited in GEO, SRA, ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), and any other domain-specific sites that are appropriate for source data deposition.
Consider middle-ground data sharing approaches for sensitive data
Raw source data is not always fully shareable with the public. This can be because of data use restrictions (see rule 4) or privacy concerns (see rule 5). Alternative mechanisms exist for sharing portions of data with the research community. For example, the database for Genotypes and Phenotypes or dbGaP (https://www.ncbi.nlm.nih.gov/gap) provides data holders with two levels of access: open and controlled. The open selection allows for broad release of nonsensitive data online, whereas the controlled release allows sensitive datasets to be shared with other investigators, provided certain restrictions are met. This increases the ability for researchers to share portions of their data that would not be shareable otherwise.
In addition to the restricted data sharing option provided by dbGaP, others have looked at ways of developing middle-ground approaches for sharing sensitive raw data or metadata. Several of these mid-level approaches use federated access systems that allow researchers to query databases containing sensitive data while preventing direct access to the data itself. An example within the United States is the Shared Health Research Information Network (SHRINE), which provides a federated system that is HIPAA-compliant.[15] International groups have also seen success in this area. BioGrid Australia (https://www.biogrid.org.au/) allows researchers to access hundreds of thousands of health records through a linked data platform where individual data holders maintain control of their data.[16] Researchers can then be provided with authorized access to certain elements within the data while restricting access to private sections of the medical data. These mid-level approaches facilitate collaboration both within the institution (i.e., across departments) and across institutions by allowing researchers to access sensitive data indirectly. They can even match patients to similar patients (for association analyses) while maintaining stringent privacy constraints.[17] Others provide summary statistics computed over large cohorts (e.g., ExAC browser/database), which maintains privacy while providing others with important information about the populations that can be used in subsequent analyses and comparisons.
Funding
MRB was supported by NLM T15 LM00707 from Jul 2014–Jun 2016 and by the NCATS, NIH, through TL1 TR000082, formerly the NCRR, TL1 RR024158 from Jul 2016–Jun 2017. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors have declared that no competing interests exist.
References
- ↑ Reichman, O.J.; Jones, M.B.; Schildhauer, M.P. (2011). "Challenges and opportunities of open data in ecology". Science 331 (6018): 703–5. doi:10.1126/science.1197962. PMID 21311007.
- ↑ Bozeman, B.; Fay, D.; Slade, C.P. (2013). "Research collaboration in universities and academic entrepreneurship: the-state-of-the-art". The Journal of Technology Transfer 38 (1): 1–67. doi:10.1007/s10961-012-9281-8.
- ↑ Brown, P.; Morello-Frosch, R.; Brody, J.G. (2008). "IRB Challenges in Multi-Partner Community-Based Participatory Research". Proceedings of The American Sociological Association Annual Meeting 2008: 1-31. https://www.brown.edu/research/research-ethics/irb-challenges-multi-partner-community-based-participatory-research.
- ↑ Vicens, Q.; Bourne, P.E. (2007). "Ten simple rules for a successful collaboration". PLOS Computational Biology 3 (3): e44. doi:10.1371/journal.pcbi.0030044. PMC PMC1847992. PMID 17397252. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1847992.
- ↑ 5.0 5.1 Jones, B.F.; Wuchty, S.; Uzzi, B. (2008). "Multi-university research teams: shifting impact, geography, and stratification in science". Science 322 (5905): 1259-62. doi:10.1126/science.1158357. PMID 18845711.
- ↑ Börner, K.; Contractor, N.; Falk-Krzesinski, H.J. et al. (2010). "A multi-level systems perspective for the science of team science". Science Translational Medicine 2 (49): 49cm24. doi:10.1126/scitranslmed.3001399. PMC PMC3527819. PMID 20844283. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3527819.
- ↑ Gottesman, O.; Kuivaniemi, H.; Tromp, G. et al. (2013). "The Electronic Medical Records and Genomics (eMERGE) Network: Past, present, and future". Genetics in Medicine 15 (10): 761-71. doi:10.1038/gim.2013.72. PMC PMC3795928. PMID 23743551. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3795928.
- ↑ Feng, Q.; Wei, W.Q.; Chung, C.P. et al. (2016). "The effect of genetic variation in PCSK9 on the LDL-cholesterol response to statin therapy". The Pharmacogenomics Journal. doi:10.1038/tpj.2016.3. PMC PMC4995153. PMID 26902539. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4995153.
- ↑ Karch, C.M.; Ezerskiy, L.A.; Bertelsen, S. et al. (2016). "Alzheimer's Disease Risk Polymorphisms Regulate Gene Expression in the ZCWPW1 and the CELF1 Loci". PLOS One 11 (2): e0148717. doi:10.1371/journal.pone.0148717. PMC PMC4769299. PMID 26919393. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4769299.
- ↑ Malik, R.; Traylor, M.; Pulit, S.L. et al. (2016). "Low-frequency and common genetic variation in ischemic stroke: The METASTROKE collaboration". Neurology 86 (13): 1217-26. doi:10.1212/WNL.0000000000002528. PMC PMC4818561. PMID 26935894. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4818561.
- ↑ Stang, P.E.; Ryan, P.B.; Racoosin, J.A. et al. (2010). "Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership". Annals of Internal Medicine 153 (9): 600–6. doi:10.7326/0003-4819-153-9-201011020-00010. PMID 21041580.
- ↑ 12.0 12.1 Prlić, A.; Procter, J.B. (2012). "Ten simple rules for the open development of scientific software". PLOS Computational Biology 8 (12): e1002802. doi:10.1371/journal.pcbi.1002802. PMC PMC3516539. PMID 23236269. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3516539.
- ↑ 13.0 13.1 Masum, H.; Rao, A.; Good, B.M. et al. (2013). "Ten simple rules for cultivating open science and collaborative R&D". PLOS Computational Biology 9 (9): e1003244. doi:10.1371/journal.pcbi.1003244. PMC PMC3784487. PMID 24086123. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3784487.
- ↑ Pearlson, G. (2009). "Multisite collaborations and large databases in psychiatric neuroimaging: Advantages, problems, and challenges". Schizophrenia Bulletin 35 (1): 1–2. doi:10.1093/schbul/sbn166. PMC PMC2643967. PMID 19023121. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2643967.
- ↑ Merriel, R.B.; Gibbs, P.; O'Brien, T.J. et al. (2011). "BioGrid Australia facilitates collaborative medical and bioinformatics research across hospitals and medical research institutes by linking data from diverse disease and data types". Human Mutation 32 (5): 517-25. doi:10.1002/humu.21437. PMID 21309032.
- ↑ Boyle, D.I.; Rafael, N. (2011). "BioGrid Australia and GRHANITE: Privacy-protecting subject matching". Studies in Health Technology and Informatics 168: 24-34. PMID 21893908.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.