Journal:Data to diagnosis in global health: A 3P approach
| Full article title | Data to diagnosis in global health: A 3P approach |
|---|---|
| Journal | BMC Medical Informatics and Decision Making |
| Author(s) | Pathinarupothi, Rahul Krishnan; Durga, P.; Rangan, Ekanath Srihari |
| Author affiliation(s) | Amrita School of Engineering, Amrita Institute of Medical Science |
| Primary contact | Email: rahulkrishnan @ am dot amrita dot edu |
| Year published | 2018 |
| Volume and issue | 18 |
| Page(s) | 78 |
| DOI | 10.1186/s12911-018-0658-y |
| ISSN | 1472-6947 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-018-0658-y |
| Download | https://bmcmedinformdecismak.biomedcentral.com/track/pdf/10.1186/s12911-018-0658-y (PDF) |
|
|
This article contains rendered mathematical formulae. You may require the Math Anywhere plugin for Chrome or the Native MathML add-on and fonts for Firefox if they don't render properly for you. |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: With connected medical devices fast becoming ubiquitous in healthcare monitoring, there is a deluge of data coming from multiple body-attached sensors. Transforming this flood of data into effective and efficient diagnosis is a major challenge.
Methods: To address this challenge, we present a "3P" approach: personalized patient monitoring, precision diagnostics, and preventive criticality alerts. In a collaborative work with doctors, we present the design, development, and testing of a healthcare data analytics and communication framework that we call RASPRO (Rapid Active Summarization for effective PROgnosis). The heart of RASPRO is "physician assist filters" (PAF) that 1. transform unwieldy multi-sensor time series data into summarized patient/disease-specific trends in steps of progressive precision as demanded by the doctor for a patient’s personalized condition, and 2. help in identifying and subsequently predictively alerting the onset of critical conditions. The output of PAFs is a clinically useful, yet extremely succinct summary of a patient’s medical condition, represented as a motif, which could be sent to remote doctors even over SMS, reducing the need for data bandwidths. We evaluate the clinical validity of these techniques using support-vector machine (SVM) learning models measuring both the predictive power and its ability to classify disease condition. We used more than 16,000 minutes of patient data (N=70) from the openly available MIMIC II database for conducting these experiments. Furthermore, we also report the clinical utility of the system through doctor feedback from a large super-speciality hospital in India.
Results: The results show that the RASPRO motifs perform as well as (and in many cases better than) raw time series data. In addition, we also see improvement in diagnostic performance using optimized sensor severity threshold ranges set using the personalization PAF severity quantizer.
Conclusion: The RASPRO-PAF system and the associated techniques are found to be useful in many healthcare applications, especially in remote patient monitoring. The personalization, precision, and prevention PAFs presented in the paper successfully shows remarkable performance in satisfying the goals of the 3Ps, thereby providing the advantages of "3As": availability, affordability, and accessibility in the global health scenario.
Keywords: precision medicine, medical informatics, personalized healthcare, motif summarization
Background
Precision medicine and personalized healthcare are quickly gaining wide research interest as well as initial acceptance among the medical community. This is facilitated by the availability of ubiquitous data sources such as wearable sensors, smartphones, and internet of things (IoT) devices, along with machine learning and large-scale data analytics tools, resulting in promising outcomes in some of the niche medical domains. Our research particularly focuses on introducing the three Ps: precision, personalization, and preventive diagnosis in remote healthcare monitoring of patients, especially in a global health scenario. In our system, patients in remote areas use wearable devices to capture their vital parameters such as blood pressure (BP), blood glucose, oxygen saturation (SpO2), electro cardiographs (ECG) etc., and transmit them to doctors in tertiary care hospitals, who in turn are expected to suggest suitably needed timely interventions. While deploying our system in the highly populous region of southern India, we found that although this promises to provide hitherto unavailable healthcare services to a critically ill and aging population, particularly in the developing world, there are significant roadblocks in our expectation that doctors embrace this new paradigm in handling patients. The doctors, who are already overloaded, feel even more overwhelmed by the voluminous data flooding in from remote patients’ sensors. Furthermore, interpreting such incoming multi-parameter data simultaneously from a multitude of remote patients is time-consuming and soon transforms into an unmanageable deluge.
Approach
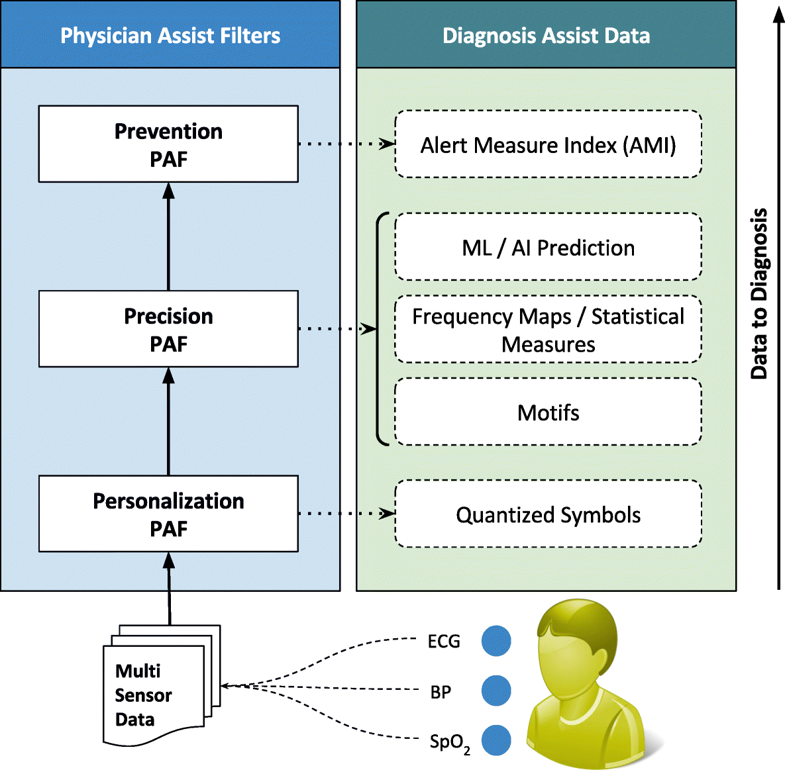
In this paper, we propose novel approaches to transform data into diagnosis. As a collaborative work between our researchers and clinicians in one of the largest super-specialty hospitals in India (Amrita Institute of Medical Sciences - AIMS), we developed physician assist filters (PAFs) that are designed to transform unwieldy time series sensor data into summarized patient/disease-specific trends in steps of progressive precision as demanded by the doctor for patient’s personalized condition at hand, and help in identifying and subsequently predictively alerting the onset of critical conditions. Together with the communication network and data transmission architecture, this new framework that we have designed, developed, and successfully deployed is called RASPRO (Rapid Active Summarization for effective PROgnosis) and was first introduced in 2016 IEEE Wireless Health.[1]
Related work
We begin by analyzing the existing systems that simply generate alerts every time one or more sensors cross the abnormality thresholds. Due to the sheer volume of such alerts, they are difficult to manage, even in the case of hospital in-patient settings, let alone for a much larger number of remotely monitored patients. Starting from some of the initial attempts reported by Anliker et al.[2], to more recent works from various researchers[3][4][5][6], the severity detection and alert generation is typically based either on predefined thresholds, or based on training of thresholds using machine learning followed by online classification of multi-sensor data. Very similar techniques of machine learning have also been used in fall detection.[7][8] Hristoskova et al.[9] propose another system wherein patient conditions are mapped to medical conditions using ontology-driven methods, and alerts are generated based on corresponding risk stratification.
Even though there has been noticeable success in detection and diagnosis of specific disease conditions, most of these works have not explored the opportunity for personalized and precision diagnosis. In an extensive review of Big Data for Health, Andreu-Perez et al.[10] specifically emphasize the opportunity for stratified patient management and personalized health diagnostics, citing examples of customized blood pressure management.[11] More specifically, Bates et al.[12] discuss the utility of using analytics to predict adverse events, which could reduce the associated morbidity and mortality rates. The authors further argue that patient data analytics based on early information supplied to the hospital prior to admission can result in better management of staffing and other hospital resources.[12] One of the recent works in personalized criticality detection is reported by Sung et al.[13], who propose an analytical unit in which the Improved Particle Swarm Optimization (IPSO) algorithm is used to arrive at patient-specific threat ranges.
To improve precision in diagnosis we also need to arrive at a balance between a completely automated system on one hand, and physician assist systems on the other. Celler et al.[14] propose a balanced approach wherein sophisticated analytics are presented to physicians, who in turn identify the changes and decide on the diagnosis. This is also supported by many results, including those reported by Skubic et al.[6], wherein domain knowledge-based methods performed as well as other trained machine learning models. These arguments and results provide further impetus for personalized, precision, and preventive diagnostic techniques that are amenable to physician interventions.
Methods
The first significant improvement that we applied is the quantization of every remotely sensed parameter based on its own customized severity boundaries. Sequential time windows of such quantized values are examined for dominant appearances of normal results or abnormalities, as the case may be, and motifs corresponding to them are extracted. Using factors set by doctors, the system then transforms these motifs by generating interventional time alerts as per clinically prescribed protocols. Both the alerts and motifs are amenable to rapid transmission to doctors, even as SMS messages on bare-minimum, bandwidth-starved wide area wireless networks. This results in the generation of more clinically relevant critical information, along with a drastic reduction in reporting every minor aberrational data that may not be indicative of any serious condition, after all. The system does not stop here. The attending doctors, when they view the alerts and/or motifs, have the luxury to request detailed data on demand (dubbed "DD-on-D"), upon which the next level of detail in the data is transmitted. This level of detail could be a straightforward frequency map of normal and abnormal values, or much more intelligent machine learning classifications in the case of proven disease conditions. The heart of our system is a framework called RASPRO (see Fig. 1), consisting of physician assist filters (PAFs) that, in going from data to diagnosis, implement the three Ps: precision, personalization, and prevention. In the following sections we describe each of these three concepts in detail.
|
Personalization PAF
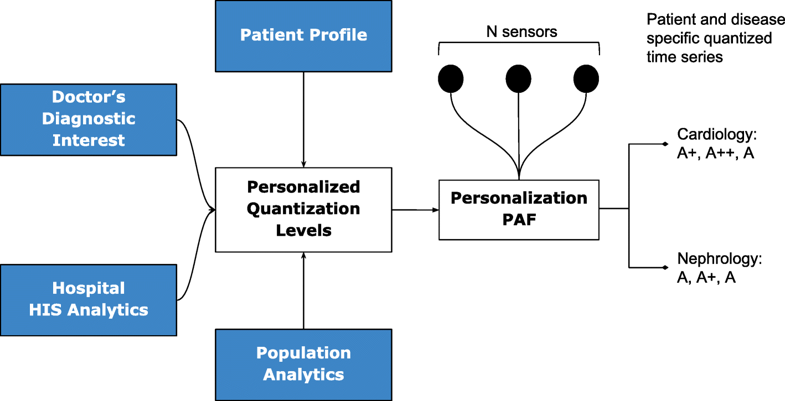
Due to the distributed data gathering and processing architecture, there is an opportunity to enhance personalization in diagnosis and treatment. The first component in the RASPRO framework, the Personalization PAF takes the form of a patient- and disease-condition-specific severity quantizer that converts raw sensor values to a series of clinically relevant severity symbols.
Adaptive qauntization
In general, let us consider N body sensors, S1,S2,…,SN with varying sensing frequencies f1,f2,…,fN. The raw time series values from these sensors are converted to discrete severity level symbols by the quantizer. The number of severity levels Li for a sensor Si can be set based on the sensor and many other factors. We assume that different vital parameter sensors have a different number of severity levels, and hence L1, say the number of severity levels for a blood pressure sensor, could be equal to five, whereas, L2 (say oxygen saturation levels) could be equal to seven. In our symbolic notation, the clinically accepted normal values are assigned the symbol "A," while above-normal values are assigned with progressive degrees of severity as "A+," "A++," etc., while that of sub-normal values are assigned "A-," "A−−," etc.; the number of “+” and “-” symbols representing degree of normal and subnormal severity respectively. Figure 2 depicts how various severity levels are arrived at in the Personalization PAF severity quantizer.
|
The quantized severity symbols are arranged into a patient-specific matrix (PSM) of N rows and W columns, where N is the total number of sensors being observed, and W is a time window in which the data is summarized. The value of W can be set by a physician or automatically derived based on the risk perception of that particular patient.
Personalization
The quantization breadth are decided by doctors based on the patient profile (or history), doctor’s diagnostic interest (for instance, a cardiologist may assign severity ranges differently from that of a nephrologist), severity ranges as suggested by using analytics on a local hospital information system (HIS), and also based on population analytics across multiple HIS spanning multiple hospitals or even from publicly available databases such as PhysioNet.[15] Together, this approach gives ample flexibility in achieving customization in inter-patient, inter-disease, intra-patient, inter-specialty diagnosis from multi-sensor data.
Precision PAF
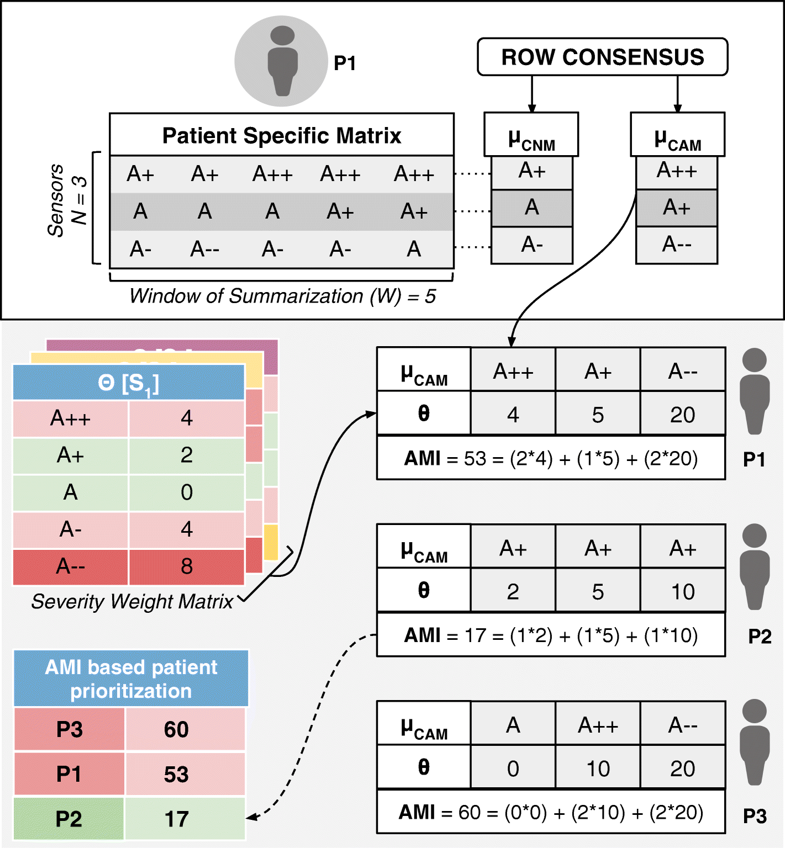
Whereas in most other applications precision directly translates into great detail in data, in remote health monitoring, precision cannot come at a cost of voluminous data presentation to the doctor. Compactness has to be retained. We have developed a step-wise refinement process for precision, which is delivered on-demand to the attending doctor. Step 1 is “Consensus Motifs (CM)”; step 2 is a collection of statistical parameters, including severity frequency maps (SFMs); and step 3 is machine learning (ML). In the first step, motifs corresponding to commonly seen normal results and abnormalities in the severity symbols series are extracted. The outcome of this is two severity summaries: (1) the most frequent trend in sensor data that we call consensus normal motif (CNM), and (2) the most frequently occurring abnormality that we term as consensus abnormality motif (CAM). The construction of this involves the following building blocks:
- Candidate symbol: α[p] is the p-th quantized severity symbol in a row of the PSM, α[1],α[2],…,α[p],…,α[W].
- Normal symbol: αNORM is a candidate symbol that represents the normal level, and its value is equal to “A” for every sensor.
- Now, let the set Cn denote all the candidate symbols in a W-long observation window, corresponding to n-th sensor in the PSM. However, we have dropped the subscript n for better clarity of discussion.
- Let σ[p] denote the sum of hamming distances of α[p] from all other candidate symbols in C such that:
- where, D(α[p],α[i]) is the hamming distance of α[p] from α[i]. Here, we assume that the hamming distance between neighboring severity levels (say, A and A+) is 1. We define a set H of all σ’s such that:
- .
- Consensus normal symbol: αCNS[C] is defined as a candidate symbol among all the symbols in C that satisfies the following two conditions: (1) its hamming distance from the normal symbol, denoted as D(αCNS[C],αNORM), is less than a sensor specific near-normal severity threshold S[n]THRESH, and (2) its sum of hamming distances from all other candidate symbols in C is the minimum. This is formulated as:
- Consensus abnormality symbol: αCAS[C] is defined as a candidate symbol in C that satisfies the following two conditions: its hamming distance from normal symbol D(αCNS[C],αNORM) is greater than or equal to a sensor specific near-normal severity thresholdS[n]THRESH and the sum of hamming distances from all other candidate symbols in C is the minimum. This is formulated as:
- Consensus normal motif: μCNM[P] is an ordered sequence of consensus normal symbols belonging to N rows in the PSM of a patient P, and is represented as <αCNS[C1],αCNS[C2],…,αCNS[CN]>. The n-th consensus normal symbol αCNS[CN] in μCNM[P] can be indexed as μCNM[P][n].
- Consensus abnormality motif: μCAM[P] is an ordered sequence of consensus abnormality symbols belonging to N rows in the PSM of patient P, which is represented as <αCAS[C1],αCAS[C2],…,αCAS[CN]>. The n-th consensus abnormality symbol αCAS[CN] in μCAM[P] can be indexed as μCAM[P][n].
- To reiterate in the above formulation, each row of a PSM is considered as an observation window set C (corresponding to a summarization time window W) to find the corresponding consensus symbols, αCNS[C] and αCAS[C]. The sequence of these symbols over the N rows in a PSM form column vector motifs μCNM[P] and μCAM[P] (refer to Fig. 3).
Fig. 3 RASPRO severity detection, summarization, and AMI calculated using CAMs and sensor specific severity weight matrix. It also shows an AMI-based patient prioritization table that can help physicians in attending to the neediest patient.
In subsequent steps of Precision PAF, the system generates a frequency map that shows how frequently different multi-sensor parameters have crossed the personalized severity thresholds. Finally, the motif time series is further used as input to proven deep learning (DL) and machine learning (ML) techniques such as Long Short Term Memory (LSTM) recurrent neural networks (RNN) [16] or Support Vector Machines (SVM) [17] that could help the doctors in diagnosis. In the next section, we use the above consensus motifs for alert generation to aid in criticality prevention.
References
- ↑ Pathinarupothi, R.K.; Rangan, E.S.; Alangot, B. et al. (2016). "RASPRO: Rapid summarization for effective prognosis in wireless remote health monitoring". 2016 IEEE Wireless Health: 1–6. doi:10.1109/WH.2016.7764566.
- ↑ Anliker, U.; Ward, J.A.; Lukowicz, P. (2004). "AMON: A wearable multiparameter medical monitoring and alert system". IEEE Transactions on Information Technology in Biomedicine 8 (4): 415–27. PMID 15615032.
- ↑ Baig, M.M.; GholamHosseini, H.; Connolly, M.J. et al. (2014). "Real-time vital signs monitoring and interpretation system for early detection of multiple physical signs in older adults". Proceeding from the IEEE-EMBS International Conference on Biomedical and Health Informatics: 355–8. doi:10.1109/BHI.2014.6864376.
- ↑ Rajevenceltha, J.; Kumar, C.S.; Kimar, A.A. (2016). "Improving the performance of multi-parameter patient monitors using feature mapping and decision fusion". Proceedings from the 2016 IEEE Region 10 Conference: 1515–8. doi:10.1109/TENCON.2016.7848268.
- ↑ Sreejith, S.; Rahul, S.; Jisha, R.C. (2016). "A Real Time Patient Monitoring System for Heart Disease Prediction Using Random Forest Algorithm". Advances in Signal Processing and Intelligent Recognition Systems 425: 485–500. doi:10.1007/978-3-319-28658-7_41.
- ↑ 6.0 6.1 Skubic, M.; Guevara, R.D.; Rantz, M. (2015). "Automated Health Alerts Using In-Home Sensor Data for Embedded Health Assessment". IEEE Journal of Translational Engineering in Health and Medicine 3: 1–11. doi:10.1109/JTEHM.2015.2421499.
- ↑ Lopes. I.C.; Vaidya, B.; Rodrigues, J.J.P.C. (2013). "Towards an autonomous fall detection and alerting system on a mobile and pervasive environment". Telecommunications Systems 52 (4): 2299–310. doi:10.1007/s11235-011-9534-0.
- ↑ Balasubramanian, A.; Wang, J.; Prabhakaran, B. (2016). "Discovering Multidimensional Motifs in Physiological Signals for Personalized Healthcare". EEE Journal of Selected Topics in Signal Processing 10 (5): 832–41. doi:10.1109/JSTSP.2016.2543679.
- ↑ Hristoskova, A.; Sakkalis, V.; Zacharioudakis, G. et al. (2014). "Ontology-driven monitoring of patient's vital signs enabling personalized medical detection and alert". Sensors 14 (1): 1598-628. doi:10.3390/s140101598. PMC PMC3926628. PMID 24445411. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3926628.
- ↑ Andreu-Perez, J.; Poon, C.C.; Merrifield, R.D. et al. (2015). "Big data for health". IEEE Journal of Biomedical and Health Informatics 19 (4): 1193-208. doi:10.1109/JBHI.2015.2450362. PMID 26173222.
- ↑ Liu, Q.; Yan, B.P.; Yu, C.M. et al. (2014). "Attenuation of systolic blood pressure and pulse transit time hysteresis during exercise and recovery in cardiovascular patients". IEEE Transactions on Bio-medical engineering 61 (2): 346–52. doi:10.1109/TBME.2013.2286998. PMID 24158470.
- ↑ 12.0 12.1 Bates, D.W.; Saria, S.; Ohno-Machado, L. et al. (2014). "Big data in health care: using analytics to identify and manage high-risk and high-cost patients". Health Affairs 33 (7): 1123-31. doi:10.1377/hlthaff.2014.0041. PMID 25006137.
- ↑ Sung, W.-T.; Chen, J.-H.; Chang, K.-W. (2014). "Mobile Physiological Measurement Platform With Cloud and Analysis Functions Implemented via IPSO". IEEE Sensors Journal 14 (1): 111–23. doi:10.1109/JSEN.2013.2280398.
- ↑ Celler, B.G.; Sparks, R.S. (2015). "Home telemonitoring of vital signs--technical challenges and future directions". IEEE Journal of Biomedical and Health Informatics 19 (1): 82–91. doi:10.1109/JBHI.2014.2351413. PMID 25163076.
- ↑ Goldberger, A.L.; Amaral, L.A.; Glass, L. (2000). "PhysioBank, PhysioToolkit, and PhysioNet". Circulation 101 (23): e215–e220. doi:10.1161/01.CIR.101.23.e215.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Grammar and punctuation was edited to American English, and in some cases additional context was added to text when necessary. In some cases important information was missing from the references, and that information was added.