Difference between revisions of "File:Fig1 Brusniak BMCBioinformatics2019 20.png"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Added summary.) |
||
| Line 1: | Line 1: | ||
== Summary == | |||
{{Information | |||
|Description='''Figure 1.''' Protein Engineering Workflow. The bioinformatics data/literature mining with or without therapeutic targets is the starting point of root protein sequences. The software allows in-silico designed protein sequences as starting points as well as those with Uniprot designations. The majority of proteins we have explored are from sequences harvested from publicly available genomes. Thus, they have species and Uniprot numbers in the Homologue sample set database fields. Black arrows show the typical engineering paths. The dotted line from high throughput production is rare due to the amount of protein produced at this scale and current lack of efficient purification protocols. Red arrows indicate going back up the hierarchy to redesign proteins based on failures or other criteria (purity, express-ability by recombinant protein expression system, synthesizability etc.) | |||
|Source={{cite journal |title= Laboratory information management software for engineered mini-protein therapeutic workflow |journal=BMC Bioinformatics |author=Brusniak, M.-Y.; Ramos, H.; Lee, B.; Olson, J.M. |volume=20 |pages=343 |year=2019 |doi=10.1186/s12859-019-2935-x}} | |||
|Author=Brusniak, M.-Y.; Ramos, H.; Lee, B.; Olson, J.M. | |||
|Date=2019 | |||
|Permission=[https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International] | |||
}} | |||
== Licensing == | == Licensing == | ||
{{cc-by-4.0}} | {{cc-by-4.0}} | ||
Latest revision as of 20:45, 24 June 2019
Summary
| Description |
Figure 1. Protein Engineering Workflow. The bioinformatics data/literature mining with or without therapeutic targets is the starting point of root protein sequences. The software allows in-silico designed protein sequences as starting points as well as those with Uniprot designations. The majority of proteins we have explored are from sequences harvested from publicly available genomes. Thus, they have species and Uniprot numbers in the Homologue sample set database fields. Black arrows show the typical engineering paths. The dotted line from high throughput production is rare due to the amount of protein produced at this scale and current lack of efficient purification protocols. Red arrows indicate going back up the hierarchy to redesign proteins based on failures or other criteria (purity, express-ability by recombinant protein expression system, synthesizability etc.) |
|---|---|
| Source |
Brusniak, M.-Y.; Ramos, H.; Lee, B.; Olson, J.M. (2019). "Laboratory information management software for engineered mini-protein therapeutic workflow". BMC Bioinformatics 20: 343. doi:10.1186/s12859-019-2935-x. |
| Date |
2019 |
| Author |
Brusniak, M.-Y.; Ramos, H.; Lee, B.; Olson, J.M. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 4.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 20:42, 24 June 2019 | 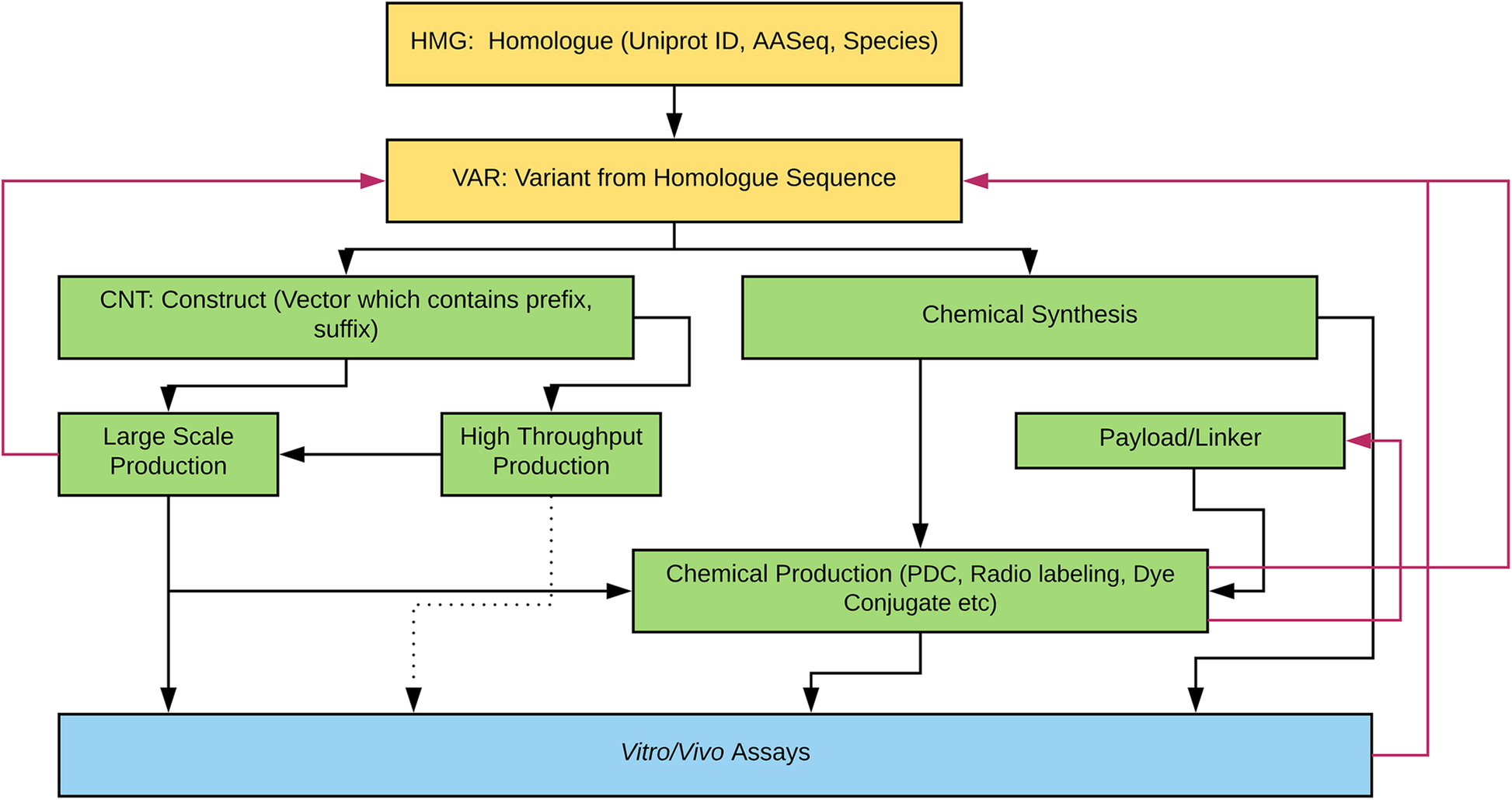 | 1,946 × 1,029 (255 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following 2 pages use this file:









