Journal:Ten simple rules for managing laboratory information
| Full article title | Ten simple rules for managing laboratory information |
|---|---|
| Journal | PLoS Computational Biology |
| Author(s) | Berezin, Casey-Tyler; Aguilera, Luis U.; Billerbeck, Sonja; Bourne, Philip E.; Densmore, Douglas; Freemont, Paul; Gorochowski, Thomas E.; Hernandez, Sarah I.; Hillson, Nathan J.; King, Connor R.; Köpke, Michael; Ma, Shuyi; Miller, Katie M.; Moon, Tae Seok; Moore, Jason H.; Munsky, Brian; Myers, Chris J.; Nicholas, Dequina A.; Peccoud, Samuel J.; Zhou, Wen; Peccoud, Jean |
| Author affiliation(s) | Colorado State University, University of Groningen, University of Virginia, Boston University, Imperial College, University of Bristol, Lawrence Berkeley National Laboratory, US Department of Energy Agile BioFoundry, US Department of Energy Joint BioEnergy Institute, LanzaTech, University of Washington Medicine, Washington University in St. Louis, Cedars-Sinai Medical Centet, University of Colorado Boulder, University of California Irvine - Irvine |
| Primary contact | Email: jean dot peccoud at colostate dot edu |
| Year published | 2023 |
| Volume and issue | 19(12) |
| Article # | e1011652 |
| DOI | 10.1371/journal.pcbi.1011652 |
| ISSN | 1553-7358 |
| Distribution license | Creative Commons CC0 1.0 Universal |
| Website | https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1011652 |
| Download | https://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1011652&type=printable (PDF) |
Abstract
Information is the cornerstone of research, from experimental data/metadata and computational processes to complex inventories of reagents and equipment. These 10 simple rules discuss best practices for leveraging laboratory information management systems (LIMS) to transform this large information load into useful scientific findings.
Keywords: laboratory information management, laboratory management, laboratory information management systems, LIMS, computational biology, mathematical modeling, transdisciplinary research
Introduction
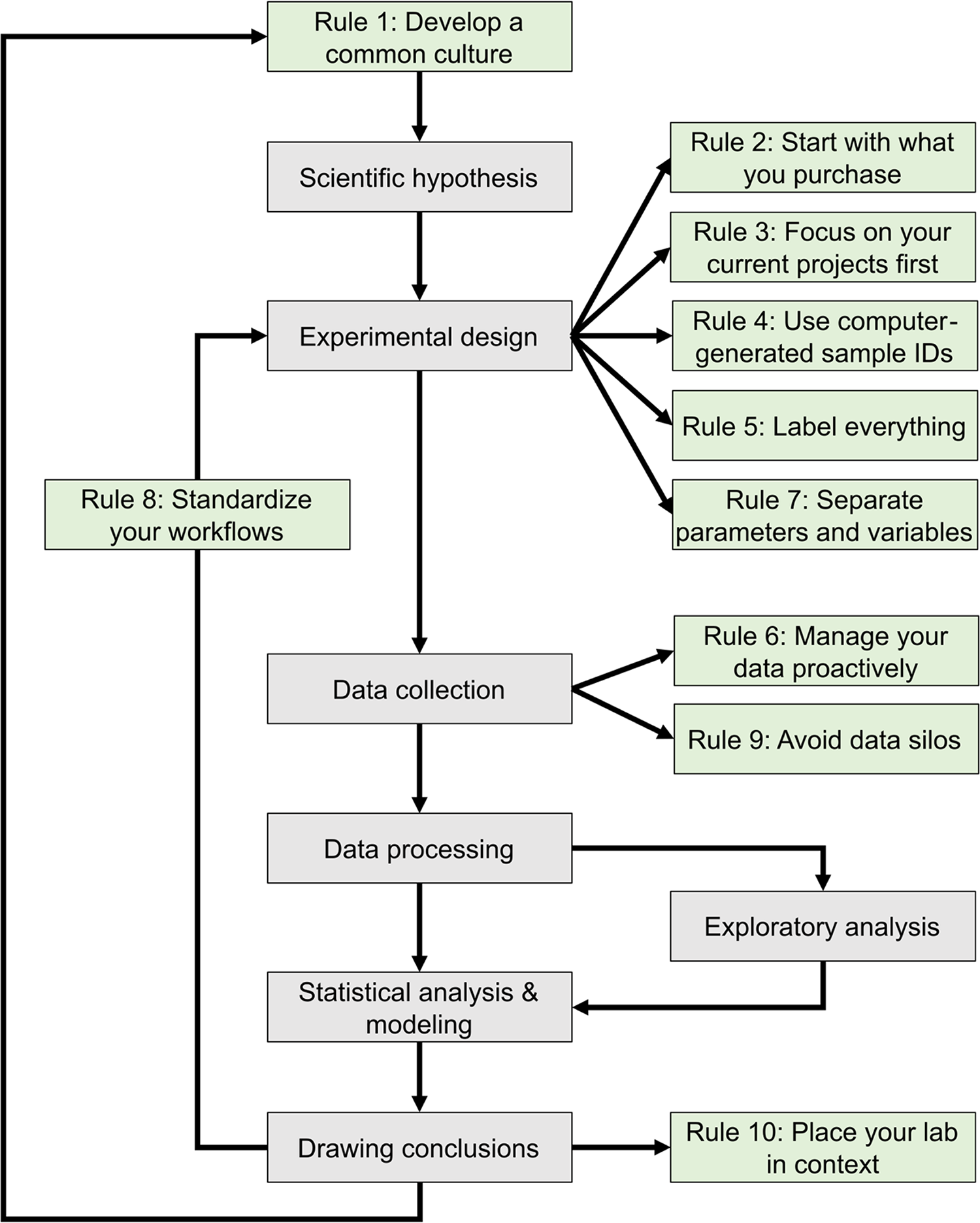
The development of mathematical models that can predict the properties of biological systems is the holy grail of computational biology. [1,2] Such models can be used to test biological hypotheses [3], quantify the risk of developing diseases [3], guide the development of biomanufactured products [4], engineer new systems meeting user-defined specifications, and much more. [4,5] Irrespective of a model’s application and the conceptual framework used to build it, the modeling process proceeds through a common iterative workflow. A model is first evaluated by fitting its parameters such that its behavior matches experimental data. Models that fit previous observations are then further validated by comparing the model predictions with the results of new observations that are outside the scope of the initial dataset (Fig. 1).
|
Historically, the collection of experimental data and the development of mathematical models were performed by different scientific communities. [6] Computational biologists had little control over the nature and quality of the data they could access. With the emergence of systems biology and synthetic biology, the boundary between experimental and computational biology has become increasingly blurred. [6] Many laboratories and junior scientists now have expertise in both producing and analyzing large volumes of digital data due to high-throughput workflows and an ever-expanding collection of digital instruments. [7] In this context, it is critically important to properly organize the exponentially growing volumes of experimental data to ensure they can support the development of models that can guide the next round of experiments. [8]
We are a group of scientists representing a broad range of scientific specialties, from clinical research to industrial biotechnology. Collectively, we have expertise in experimental biology, data science, and mathematical modeling. Some of us work in academia, while others work in industry. We have all faced the challenges of keeping track of laboratory operations to produce high-quality data suitable for analysis. We have experience using a variety of tools, including spreadsheets, open-source software, homegrown databases, and commercial solutions to manage our data. Irreproducible experiments, projects that failed to meet their goals, datasets we collected but never managed to analyze, and freezers full of unusable samples have taught us the hard way lessons that have led to these 10 simple rules for managing laboratory information.
This journal has published several sets of rules regarding best practices in overall research design [9,10], as well as the computational parts of research workflows, including data management [11–13] and software development practices. [14–16] The purpose of these 10 rules (Fig. 1) is to guide the development and configuration of laboratory information management systems (LIMS). LIMS typically offer electronic laboratory notebook (ELN), inventory, workflow planning, and data management features, allowing users to connect data production and data analysis to ensure that useful information can be extracted from experimental data and increase reproducibility. [17,18] These rules can also be used to develop training programs and lab management policies. Although we all agree that applying these rules increases the value of the data we produce in our laboratories, we also acknowledge that enforcing them is challenging. It relies on the successful integration of effective software tools, training programs, lab management policies, and the will to abide by these policies. Each lab must find the most effective way to adopt these rules to suit their unique environment.
Rule 1: Develop a common culture
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. To more easily differentiate footnotes from references, the original footnotes (which were numbered) were updated to use lowercase letters. Most footnotes referencing web pages were turned into proper citations.