Journal:SODAR: Managing multiomics study data and metadata
| Full article title | SODAR: Managing multiomics study data and metadata |
|---|---|
| Journal | GigaScience |
| Author(s) | Nieminen, Mikko; Stolpe, Oliver; Kuhring, Mathias; Weiner III, January; Pett, Patrick; Beule, Dieter; Holtgrewe, Manual |
| Author affiliation(s) | Berlin Institute of Health at Charité–Universitätsmedizin Berlin |
| Primary contact | Email: mikko dot nieminen at bih dash charite dot de |
| Year published | 2023 |
| Volume and issue | 12 |
| Article # | giad052 |
| DOI | 10.1093/gigascience/giad052 |
| ISSN | 2047-217X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giad052/7232111 |
| Download | https://academic.oup.com/gigascience/article-pdf/doi/10.1093/gigascience/giad052/50974561/giad052.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Scientists employing omics in life science studies face challenges such as the modeling of multiassay studies, recording of all relevant parameters, and managing many samples with their metadata. They must manage many large files that are the results of the assays or subsequent computation. Users with diverse backgrounds, ranging from computational scientists to wet-lab scientists, have dissimilar needs when it comes to data access, with programmatic interfaces being favored by the former and graphical ones by the latter.
We introduce SODAR, the system for omics data access and retrieval. SODAR is a software package that addresses these challenges by providing a web-based graphical user interface (GUI) for managing multiassay studies and describing them using the ISA (Investigation, Study, Assay) data model and the ISA-Tab file format. Data storage is handled using the iRODS data management system, which handles large quantities of files and substantial amounts of data. SODAR also offers programmable application programming interfaces (APIs) and command-line access for metadata and file storage.
SODAR supports complex omics integration studies and can be easily installed. The software is written in Python 3 and freely available at https://github.com/bihealth/sodar-server under the MIT license.
Keywords: omics, research, research data, data management
Introduction
Modern studies in life sciences rely on “omics” assays, which encompass branches of science such as genomics, proteomics, and metabolomics. One or multiple assays can be run within a single study, potentially including assays for multiple omics studies of several types.
The following key steps are required for executing these complex omics studies: (i) planning, which results in study metadata; (ii) collection of mass data; and (iii) data analysis, including the integration of multiple assays. The aim of SODAR is to ensure support for scientists within all the steps.
Challenges
Each step presents its own set of challenges. During planning, it is important to enable recording crucial factors and covariates. The flow of materials and samples through processes must also be specified in sufficient detail. Further challenges arise from, for example, assays using complex multiplexing, such as the need for reference samples; requirements for using controlled vocabularies or ontologies; and possible change of assays over time.
In the data collection step, scientists must record the used machines, kits, and versions of both hardware and software used. Omics studies also create large volumes of data, ranging from a few gigabytes for mass spectrometry (MS) to terabytes for imaging such as microscopy. These data may be spread among many files, further complicating the needs for managing mass data storage. Instead of a rigid process, data collection should also be adjustable to changes and developments in data generation over time.
Data analysis is often split into multiple phases, with primary analysis of each assay followed by steps for integration of results. Specific results need to be fed back to metadata management, annotation, quality control, or storing resulting markers. Access to metadata with recorded factors and confounders is necessary in each step, while access to primary raw data becomes less important after the primary analysis. Certain analysis results are written back into the mass data storage. This includes binary alignment map (BAM) files and variant call format (VCF) files.
There are also overarching challenges for the steps in study execution. All data should be recorded in structured format. Automation should be applied where possible, and on-premises installation might be preferable or even required when data privacy–relevant data are generated such as with DNA sequencing.
Data management approaches
In this and the following section, we will discuss the topic of |data management and software. The terms “data” and “document” will be used interchangeably in this section. The steps described in the prior section can be interpreted as processes taking documents and materials as input, as well as generating more documents and materials as the result. For example, data collection takes the plan document and samples and generates assay result files (documents). Scientists thus need computational tools for supporting them in managing their scientific and research data.
Historically, such documents are maintained on paper in laboratory notebooks, or documentation created by quality management systems (QMSs). For the most direct and unstructured approaches in maintaining digital data, this corresponds to word processing, spreadsheet, and image files on local or network drives. More structured approaches are desirable for taking advantage of digital documents, preventing research data loss [1] or fostering reuse. [2]
While data management in science is a broad topic, the library and information science community is frequently approaching it using a top-down approach. Frequently, in this context, the term “research data management” (RDM) is used. Here, the needs of whole organizations and their parts for managing their research data, as well as the necessary steps to establish whole RDM systems, are considered first (cf. Donner [3]). This correlates with the role of libraries in certain academic organizations for organizing data that were collected in research.
A second approach, which can be described as “bottom-up,” originates from different “working scientist” communities. The communities commonly refer to the topic as “scientific data management” (SDM) and solve their problems at hand, often starting with specific small-scale solutions, which are then upscaled if the need arises. While considering their organizational embedding, they focus on solving specific data management challenges for themselves and their peers. We found ourselves in this situation and will thus focus on this perspective.
Data management software packages
SDM needs come in different forms and shapes. We could find no general treatment of the subject of data management in the literature. Machina and Wild [4] provide a collection of four tool categories: laboratory information management systems (LIMSs), electronic laboratory notebooks (ELNs), scientific data management systems (SDMSs), and chromatography data systems (CDSs) that we generalize as instrument-specific data systems (IDS). In this section, we provide our take on explaining what these systems comprise. We also note—as Machina and Wild [4] did—that categorization of such software solutions is not clear-cut, and features may be overlapping. We expand this list by two more system types: data repository systems (DRSs) and database/data warehouse management frameworks (DMFs).
The four tools described by Machina and Wild [4] are as follows:
- A LIMS focuses on storing information around laboratory workflows. This includes tracking of consumables, samples, instruments, and tests. A LIMS deals with daily tasks of laboratories such as billing and instrument calibration. It is often specific to certain domain areas such as sequencing facilities.
- The ELN focuses on allowing humans to record their laboratory work. It replaces paper notebooks and captures experiments and their results, mostly in free-form text, pictures, tables, and so on. The ELN plays a key role in fulfilling regulatory requirements.
- The CDS/IDS provides data capturing, storage, and analysis functionality in instrument-specific domains. Two examples are the CASAVA pipeline and the BaseSpace cloud-based service, both from Illumina. The former is provided without extra cost with the instrument along with its source code, while the latter is purchasable and closed source. Such software often ships with the instruments themselves.
- The SDMS provides scientific content management functionality for scientific data and documentation. It allows for the management of metadata and potentially mass data. The SDMS's core functionality generally doesn't include data analysis, user-centric data collection, or laboratory workflow tracking. Such features may be potentially supported by plugins or extensions. Many such systems offer integration with surrounding systems (e.g., via application programming interfaces [APIs]).
We augment this list by two system types:
- The DRS provides shared access to data with appropriate documentation and metadata. Examples are FAIRdom Seek [5], Dataverse [6], and Yoda. [7] There are also specialized DRSs focusing on particular use cases, such as dbGAP [8], MetaboLights [9], and Gene Expression Omnibus [10], that allow for managing public or controlled public access to large research data collections.
- The DMF allows for the rapid development of database and data warehouse applications. It often provides preexisting components to build on ready-made functionality and extension by implementing custom components. This enables the creation of domain-specific databases and structured data capturing. Examples include Molgenis [11] and Zendro. [12]
Other types of systems also exist, and not every system falls into just one category. A complete review of such systems is beyond the scope of this article. This section identifies focus areas of systems involved in some form of scientific data management. SODAR falls into the category of SDMS.
Data management technologies
For planning and documenting experiments and their structure, experiment-oriented metadata storage formats with predefined syntax and semantics exist. A popular standard is the ISA (Investigation, Study, Assay) model [13], which allows describing studies with multiple samples and assays. The ISA model defines the ISA-Tab tabular file format, which allows users to model each processing step with each intermediate result and annotate each of these with arbitrary metadata. An example of an alternative to ISA-Tab is Portable Encapsulated Projects (PEPs). [14] There are also more specialized standards such as Brain Imaging Data Structure (BIDS) for brain imaging data [15], as well as other approaches such as Clinical Data Interchange Standards Consortium (CDISC) standards [16] and the Hierarchical Data Format (HDF5). [17] Use of generic file formats such as HDF5, TSV, XML, and JSON is also common.
For storing large volumes of omics data, it is possible to simply use file systems or object storage systems. More advanced solutions such as Shock [18] or dCache [19] allow for storing metadata and distributing data over multiple servers. iRODS (Integrated Rule-Oriented Data System) [20, 21] adds further features, such as running rules and programs within the data system and enabling integration with arbitrary authentication methods.
For publication, raw and processed data and metadata are deposited in scientific catalogs, study databases, and registries. Examples include the BioSamples database for metadata [22] and Sequence Read Archive (SRA) for raw sequencing data. [23]
Our work
In our work, we focus on managing many omics projects of varying data size and various use cases, including cancer and functional genomics studies. We also need to support multiple technologies such as whole genome sequencing, single-cell sequencing, proteomics, and MS. Deposition to public repositories was not a necessity in our context. However, SODAR is an ISA-compliant system. Should the data owner require it, it is easily feasible to create appropriate exports to public data repositories using the APIs provided by SODAR. Open-source software is a requirement to avoid vendor lock-in and allow for flexibility in different use cases. A suitable end-to-end solution was not available when we started our work in 2016. Therefore, we set out to implement an integrated system for managing omics-specific data and metadata.
In this article, we introduce SODAR (System for Omics Data Access and Retrieval). SODAR combines the modeling of studies and assays using the ISA-Tab format with handling of mass data storage using iRODS. More example projects are available in the SODAR online demo server. [41]
Results
We present the results by first giving an overview of the developed SODAR system. Next, we compare it to a selection of existing tools and their relevant features. We then describe processes we have established around SODAR. Finally, internal usage statistics are detailed along with discussion on the limitations of SODAR.
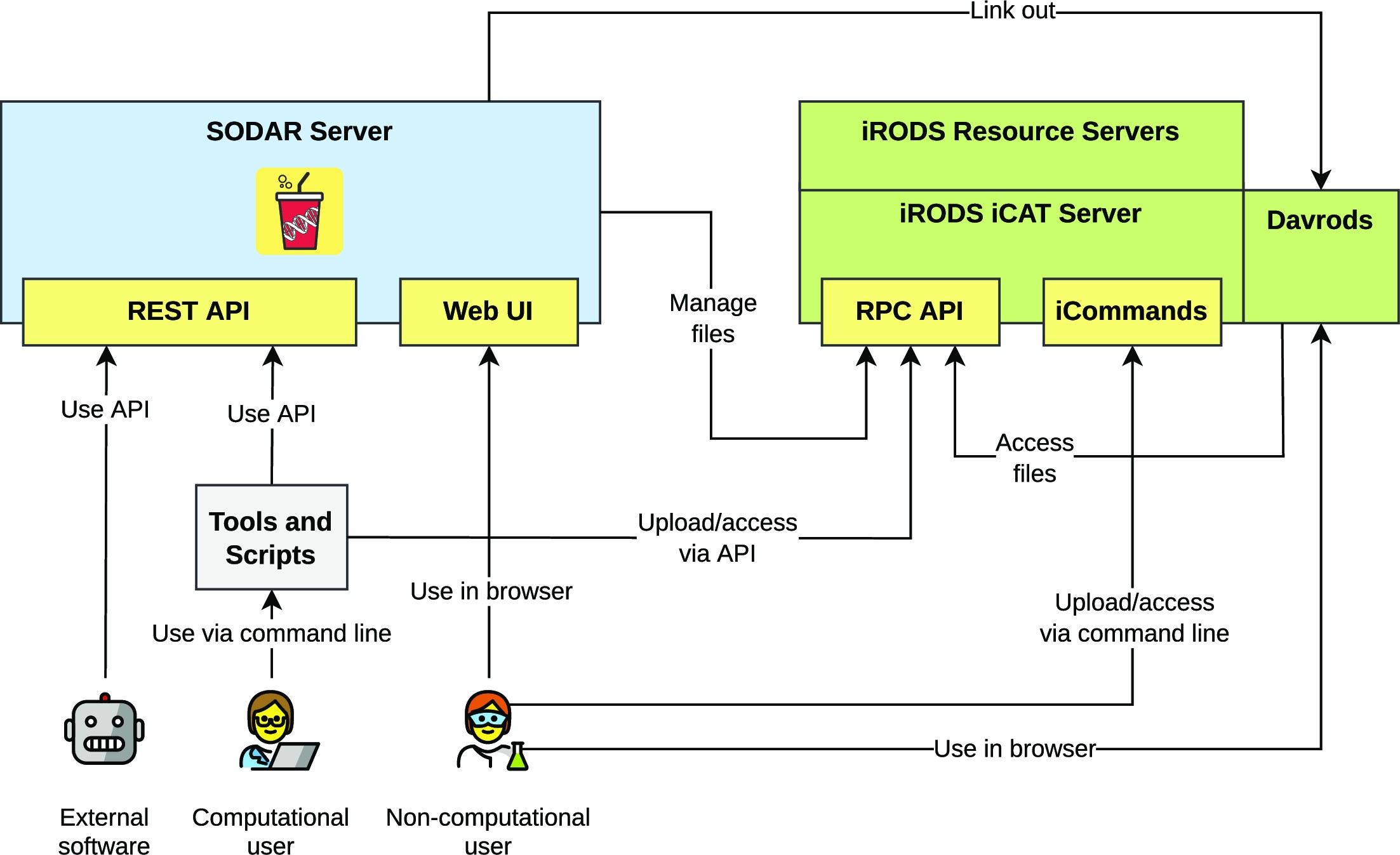
Resulting system overview
Figure 1 presents the components of the SODAR system. The SODAR server is built on the Django web framework. It contains the main system logic and provides both a graphical user interface (GUI) and APIs for managing projects, studies, and data.
|
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, grammar, and punctuation. In some cases important information was missing from the references, and that information was added.