Journal:ApE, A Plasmid Editor: A freely available DNA manipulation and visualization program
| Full article title | ApE, A Plasmid Editor: A freely available DNA manipulation and visualization program |
|---|---|
| Journal | Frontiers in Bioinformatics |
| Author(s) | Davis, M. Wayne; Jorgensen, Erik M. |
| Author affiliation(s) | Howard Hughes Medical Institute and School of Biological Sciences, University of Utah |
| Primary contact | Email: jorgensen at biology dot utah dot edu |
| Editors | Machiraju, Raghu |
| Year published | 2022 |
| Volume and issue | 40 |
| Article # | e55 |
| DOI | 10.1093/nar/gkr1288 |
| ISSN | 2673-7647 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fbinf.2022.818619/full |
| Download | https://www.frontiersin.org/articles/10.3389/fbinf.2022.818619/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
A Plasmid Editor (ApE) is a free, multi-platform application for visualizing, designing, and presenting biologically relevant DNA sequences. ApE provides a flexible framework for annotating a sequence manually or using a user-defined library of features. ApE can be used in designing plasmids and other constructs via in silico simulation of cloning methods such as polymerase chain reaction (PCR), Gibson assembly, restriction-ligation assembly, and Golden Gate assembly. In addition, ApE provides a platform for creating visually appealing linear and circular plasmid maps. It is available for Mac, PC, and Linux-based platforms and can be downloaded at https://jorgensen.biology.utah.edu/wayned/ape/.
Keywords: plasmid editor, DNA visualization, molecular biology tools, molecular techniques simulator, freely available software
Introduction
DNA visualization software must 1) annotate features and depict DNA features graphically, 2) simulate molecular cloning techniques, and 3) generate visually appealing output for figures. Good DNA visualization software applies meaning to a string of DNA bases. Fundamentally, this requires flexible annotation—applying names to a region, and visualization of functional regions—applying pictures to show the spatial relationships between sequence regions. Every piece of the DNA should be annotated with its biologically relevant attributes. In addition, a biologist must be able to identify subsequences such as restriction enzyme recognition sequences, recombinase recognition sequences, and overlapping end sequences that are useful for particular recombinant techniques.
Good DNA software also provides powerful in silico simulation of common DNA manipulations, such as restriction digests or Gibson cloning. By manipulating DNA in silico, a biologist can ensure that recombinant constructs include functionally complete pieces that have the DNA in-order and in-frame. In other words, good software allows a researcher to synthesize a working plan. This might be working backwards in silico from a desired product to determine the needed inputs. Conversely, it allows a researcher to start with a given set of available plasmids and work in the virtual laboratory to generate possible products. Finally, visualization software can be invaluable for determining whether an analytic result—a DNA sequence, a diagnostic polymerase chain reaction (PCR) or restriction digest—has generated the expected product. The scientist can use the software to align sequences or simulate gels of each step to confirm their work.
Finally, good DNA software can generate visually pleasing output with a flexible level of detail. This representation should be easily exported in an open and widely used text or graphic format. For example, text output can be used to generate class reports, student theses, or manuscripts for publication. Similarly, graphical output can be used to generate meeting posters or slides for class reports or conference presentations.
Because of this critical need for visualization software, many DNA visualization programs have been written. Many of these are written by researchers themselves to solve their own needs in the lab. Among these are Serial Cloner [Perez, 2021; AcaClone, 2021; GenBeans, 2016; York, 2021] and DNA Strider. [Douglas, 1995] Such solutions are often very powerful at solving a specific task, but they can be lacking in broad application. Similarly, they are often dependent on a single operating system, and they can sometimes have limited visual appeal in the graphic outputs. On the other hand, they are usually freely available, and as such are very accessible to small groups and teaching labs.
At the other extreme, commercial ventures have written very powerful and flexible sequence visualization packages. Popular packages include Benchling [Benchling, 2021], SnapGene [SnapGene, 2021], and Gene Construction Kit. [Gene Construction Kit, 2021] In order to have a wide customer base, they endeavor to have a complete set of analysis procedures and in silico reaction simulations. Because the visual output is usually a major factor in the product literature, the software has been carefully designed to generate visually appealing output. All of this engineering takes programmer and designer time; as such, these packages are often cost prohibitive for individual laboratories, and almost always are out-of-range of a teaching laboratory.
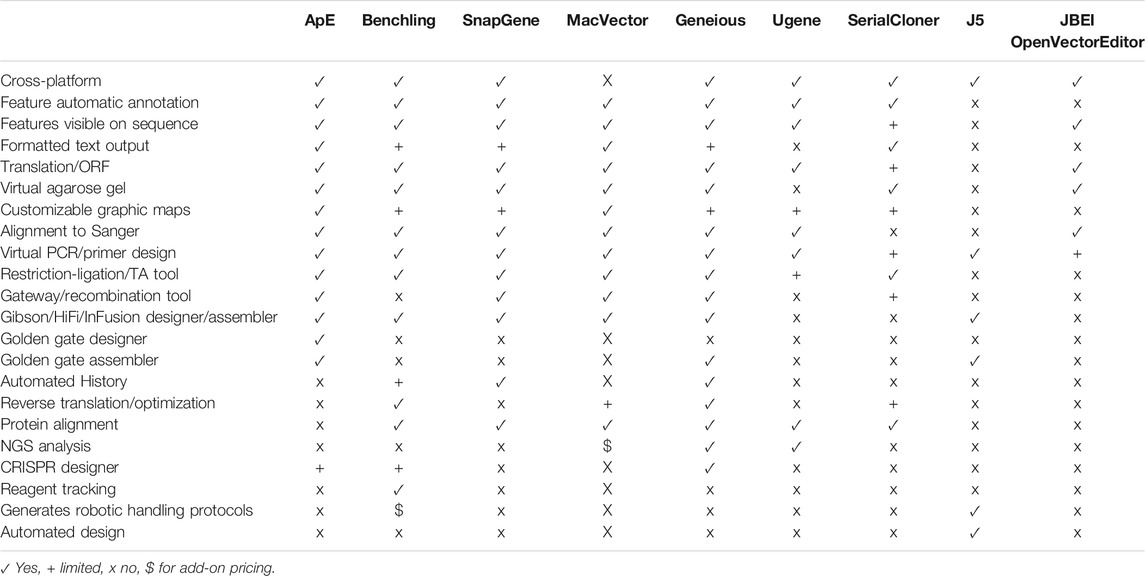
We have taken the long view to solving this problem. ApE is a freely available program written over the last 17 years by a molecular biologist for molecular biologists. Thus, it leverages the insider knowledge of what makes a successful DNA editing program. Further, the long-timeframe approach has allowed the program to become both highly versatile and streamlined; ApE now rivals the commercially available packages in both its diversity of features and its visual outputs. Importantly, unlike commercial packages, its free availability makes it well-suited for use in small labs or teaching labs. A summary of some of the features in ApE and a selected set of other visualization programs is provided in Table 1.
|
Method (code description)
Language and supported operating systems
ApE is written in Tcl/Tk. Current distribution of ApE is with Tcl/Tk version 8.6.11. [Walzer et al.] There are ready-to-run versions of ApE for Windows, MacOS, and Linux systems.
For Windows, the program is packaged into a self-contained tclkit [Wippler, 2021] using the Starkit Developer eXtension (sdx). [Thoyts, 2021] The Tclkit is a compiled binary generated by Ashok P. Nadkarni and contains the Tcl Windows API extension package (TWAPI). [Nadkarni, 2021] The .exe file was edited using Resource Hacker [Johnson, 2021] to contain a custom icon set and relevant version and copyright information. Bundled in the virtual filesystem of the .exe file are copies of the ApE accessory files (see below). The .exe is compiled as an x86-32-bit application, and the software should run on versions of Windows between Windows '98 and Windows 10.
For MacOS, ApE is packaged as an application bundle. The executable files in the bundle were generated from Tcl and Tk source. [Walzer, 2021] The current release is targeted to x86 architectures with OS versions 10.11 and above. The executable application bundle includes embedded Tcl and Tk frameworks, the Tcl script, copies of the ApE accessory files, a custom application icon, and a MacOS property list file.
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.