Journal:PIKAChU: A Python-based informatics kit for analyzing chemical units
| Full article title | PIKAChU: A Python-based informatics kit for analyzing chemical units |
|---|---|
| Journal | Journal of Cheminformatics |
| Author(s) | Terlouw, Barbara R.; Vromans, Sophie P.J.M.; Medema, Marnix H. |
| Author affiliation(s) | Wageningen University |
| Primary contact | barbara dot terlouw at wur dot nl |
| Year published | 2022 |
| Volume and issue | 14 |
| Page(s) | 34 |
| DOI | 10.1186/s13321-022-00616-5 |
| ISSN | 1758-2946 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://jcheminf.biomedcentral.com/articles/10.1186/s13321-022-00616-5 |
| Download | https://jcheminf.biomedcentral.com/track/pdf/10.1186/s13321-022-00616-5.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
As efforts to computationally describe and simulate the biochemical world become more commonplace, computer programs that are capable of in silico chemistry play an increasingly important role in biochemical research. While such programs exist, they are often dependency-heavy, difficult to navigate, or not written in Python, the programming language of choice for bioinformaticians.
Here, we introduce PIKAChU (Python-based Informatics Kit for Analysing CHemical Units), a cheminformatics toolbox with few dependencies implemented in Python. PIKAChU builds comprehensive molecular graphs from SMILES strings, which allow for easy downstream analysis and visualization of molecules. While the molecular graphs (Graphical Abstract, below) PIKAChU generates are extensive—storing and inferring information on aromaticity, chirality, charge, hybridization, and electron orbitals—PIKAChU limits itself to applications that will be sufficient for most casual users and downstream Python-based tools and databases, such as Morgan fingerprinting, similarity scoring, substructure matching, and customizable visualization. In addition, it comes with a set of functions that assists in the easy implementation of reaction mechanisms. Its minimalistic design makes PIKAChU straightforward to use and install, in stark contrast to many existing toolkits, which are more difficult to navigate and come with a plethora of dependencies that may cause compatibility issues with downstream tools. As such, PIKAChU provides an alternative for researchers for whom basic cheminformatic processing suffices, and can be easily integrated into downstream bioinformatics and cheminformatics tools. (PIKAChU is available at https://github.com/BTheDragonMaster/pikachu.)
Keywords: cheminformatics kit, Python, structure visualization, in silico chemistry, molecular fingerprinting
Introduction
In a data-driven world where the discovery of novel natural and synthetic molecules is increasingly necessary, in silico chemical processing has become an essential part of biological and chemical research. Novel metabolites are compared or added to searchable chemical databases such as ChEBI [6], PubChem [10], NP Atlas [20], and COCONUT [17]; molecular structures are predicted from biological pathways [3, 16]; and bioactivities and pharmaceutical properties are predicted from chemical structure. [1, 18, 21] Such analyses rely on robust cheminformatics kits that can perform basic chemical processing, such as fingerprint-based similarity searches, substructure matching, molecule visualization, and chemical featurization for machine learning purposes.
Typically, molecular processing by cheminformatics kits begins with the reading in of molecular data from chemical data formats, ranging from one-dimensional to three-dimensional molecular representations. One such format is the SMILES (Simplified Molecular-Input Line Entry System) format, which represents a molecule as a one-dimensional string, describing atom composition, connectivity, stereochemistry, and charge. More elaborate formats such as PDB and MOL use text files to store not just the abovementioned properties but also atom coordinates in three-dimensional space.
Depending on the application, different formats and subsequent processing are appropriate. Due to the vast number of possible chemical analyses, exhaustive cheminformatics kits have accumulated into software libraries that are so large that they can be both hard to navigate and rely on so many dependencies that they can be difficult to implement in software packages. As a result, the trade-off between time spent accessing and integrating these cheminformatics kits into a codebase and time spent on actual analyses is disproportionate for users who need to perform simple in silico analyses such as reading in SMILES, drawing a molecule, or visualizing a substructure. One popular open-source cheminformatics kit that suffers from this problem is RDKit. [11] While RDKit is an incredibly fast and powerful library that supports an immense variety of possible chemical operations, its use of both Python and C++ as programming languages, as well as the sheer number of dependencies it relies on, frequently causes compatibility issues when integrating RDKit into other programs, and disproportionately increases the number of libraries that need to be installed. Therefore, while RDKit is great for heavy-duty in silico analyses such as computing 3D conformers for a compound or constructing electron density maps, it is a bit too much for the basic operations that most researchers in bioinformatics and cheminformatics require.
A second widely-used cheminformatics kit is CDK. [22] Written in Java, it is well-suited for implementation in web applications, and it has successfully been used for molecular processing in the COCONUT database [17], the Cytoscape application chemViz2 [13], and the scientific workflow platform KNIME (Konstanz Information Miner). [2] However, with Python becoming the programming language of choice for many scientists [4], especially those working in the growing field of (deep) neural networks, CDK is not always an ideal fit.
To make basic cheminformatics processing more accessible for Python programmers, we therefore introduce PIKAChU: a Python-based Informatics Kit for Analysing Chemical Units. PIKAChU is a flexible cheminformatics tool with few dependencies. It can parse molecules from SMILES, visualize chemical structures and substructures in matplotlib, perform extended connectivity fingerprinting (ECFP) [15] and Tanimoto similarity searches, and execute basic reactions with a focus on natural product chemistry. Therefore, we hope that PIKAChU can provide a convenient alternative for many Python-based bio- and cheminformatics tools and databases that only demand basic chemical processing.
Methods and implementations
Software description
PIKAChU is implemented in Python (v3.9.7). Its only dependency is the common Python package matplotlib (v3.4.3). PIKAChU can be run on Windows, MacOS, and Linux systems.
Parsing molecules from SMILES
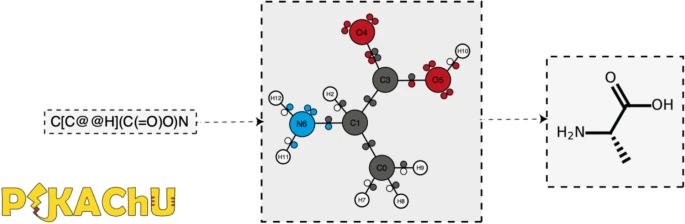
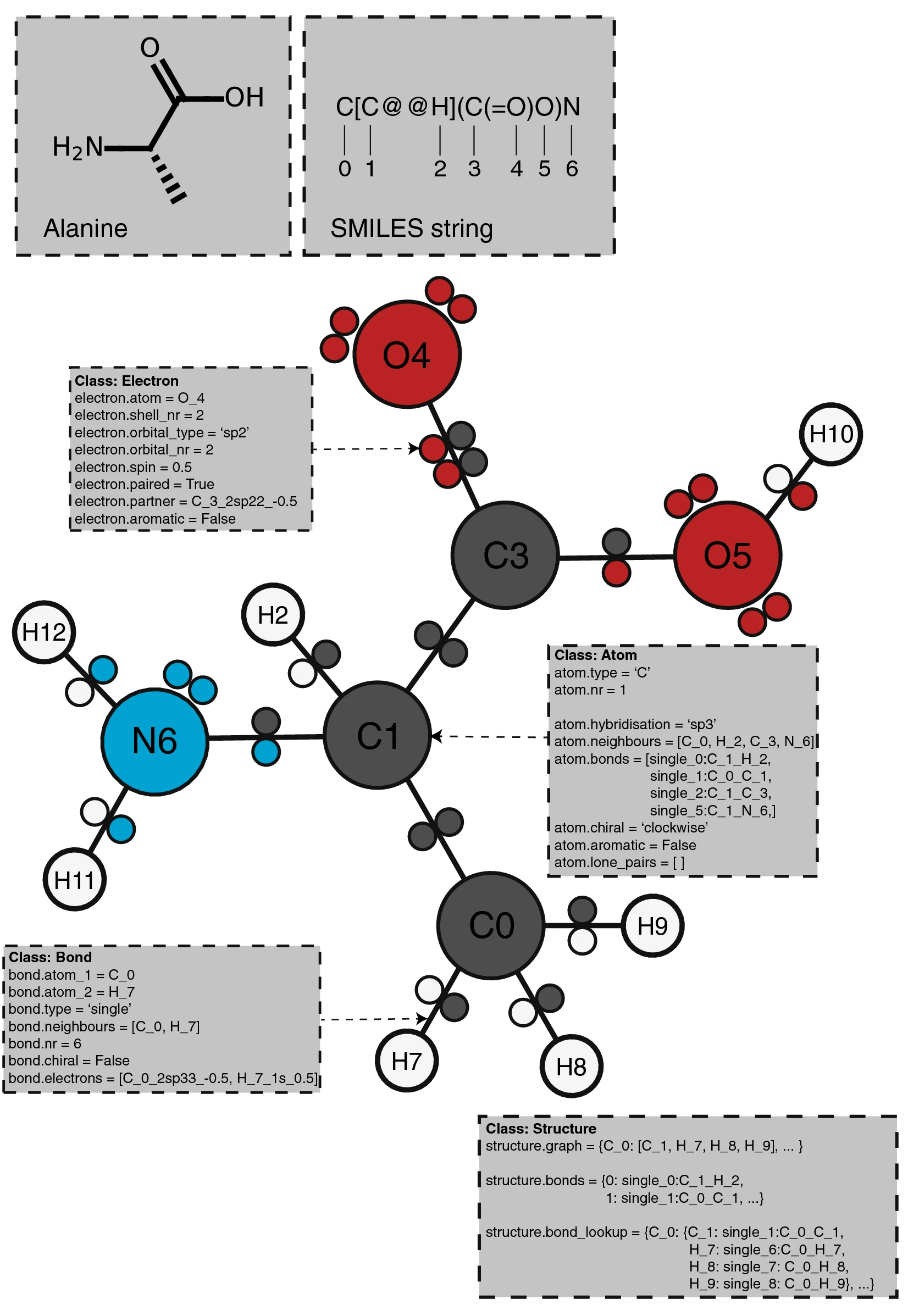
PIKAChU takes a SMILES string as input and from it builds a graph object, in which nodes represent atoms and edges represent bonds (Fig. 1). For each atom, PIKAChU initially stores information on chirality, aromaticity, charge, and connectivity. For each bond, it stores bond type (single, double, triple, quadruple, or aromatic), neighboring atoms, and cis–trans stereochemistry for double bonds. Once all atoms, bonds, and their connections have been stored, electron shells and orbitals are constructed for each non-hydrogen atom. Next, we determine the valence for each non-hydrogen atom, taking into account atom charge. For atoms of variable valence such as sulfur (2, 4 or 6) and phosphorus (3 or 5), we select a valence that is equal to or higher than the sum of non-hydrogen bonds and explicit hydrogen bonds, prioritizing smaller valences. Double, triple, quadruple, and aromatic bonds contribute proportionally to this sum. If insufficient bonding orbitals are available to achieve the desired valence, the electrons in the valence shell are excited to higher-energy orbitals, such that each orbital contains at most one electron. Implicit hydrogens are then added to the structure such that the pre-determined valences are obeyed.
|
An exception is made for nitrogen with valence 5, which is not chemically possible due to insufficient bonding orbitals but can sometimes be encountered in SMILES strings, especially in those describing compounds containing nitro groups. If such a valence 5 nitrogen is attached to at least one oxygen through a double bond, this double bond is interpreted as a single bond instead, the oxygen’s charge is set to − 1 and the nitrogen’s charge is set to 1, such that the nitrogen’s valence becomes 4 and bonding laws are obeyed.
Subsequently, electrons are allocated to the p-orbitals of π bonds in double, triple, and quadruple bonds, and atom hybridization is determined from steric number. Then, all cycles in the graph are detected using an open-source Python implementation [12] of the simple cycle detection algorithm described by Johnson in 1975. [8] PIKAChU removes all cycles smaller than three atoms and identifies the smallest set of unique smallest rings (SSSR).
Next, the SSSR is used for aromaticity detection. This is done recursively: in each round, each cycle that has not yet been added to the set of aromatic cycles is evaluated with Hückel’s 4n + 2 rule on planar rings. We chose to assess aromatic cycles rather than systems; Hückel’s rule is not always reliable for cyclic systems. [7] First, the hybridization of all atoms in the cycle is examined. All atoms must be sp2-hybridised, or sp3-hybridised with a delocalizable lone pair that can be promoted to a p-orbital. If the cycle is planar and the sum of double bonds and lone pairs is odd, the cycle is considered aromatic. Aromatic bond stretches are locally kekulized, and double bonds are subsequently counted. When a cycle is considered aromatic, bonds and atoms in the cycle are set to aromatic, and lone pairs of sp3-hybridised atoms are promoted to p-orbitals such that the new hybridization is sp2. Recursion is needed in case double bonds in cyclic systems are defined in such a way that not all sub-cycles contain the required number of bonds to obey Hückel’s rule: when adjacent bonds are updated to aromatic, they will be counted in the next round of aromaticity detection (Additional file 2: Fig. S1). When, after an iteration, the number of aromatic cycles no longer changes, all aromatic cycles have been detected. From these cycles, PIKAChU defines aromatic systems, where aromatic cycles are considered part of an aromatic system if they share a bond with at least one other aromatic cycle in the system.
Electrons involved in σ bonds and aromatic bonds are only allocated after aromaticity detection. As electrons involved in aromatic systems are not localized to specific atoms or bonds, the p-orbitals of atoms in aromatic systems are emptied and their electrons stored in an AromaticSystem object.
Finally, any unpaired electrons are dropped back to lower-energy orbitals. A structure object is returned which can be visualized, kekulized, analyzed through substructure matching and molecular fingerprinting, and altered through an assortment of built-in and custom chemical reactions.
If a SMILES string yields a structure object that is chemically incorrect due to too many or too few bonds being attached to an atom or valence shells not being filled appropriately in the case of organic atoms, PIKAChU gives a StructureError, informing the user that the parsed structure is chemically incorrect and gives a rough indication of why. Two examples of such StructureError messages are "Error parsing 'F/C(\Cl)=C(F)/Cl': Conflicting double bond stereochemistry" and "Error parsing 'CN(=O)=O': Basic bonding laws have been violated."
Visualization and kekulization
Prior to visualization, aromatic systems within a structure are kekulized so that aromatic systems can be represented by alternating single and double bonds. PIKAChU kekulizes aromatic systems using a Python implementation [23] of Edmonds’ Blossom Algorithm for maximum matching. [5] Next, atoms are positioned using PIKAChU’s drawing software. PIKAChU’s Python-based drawing algorithm was adapted and improved from SmilesDrawer [14], an open-source JavaScript library for molecular visualization. While written in different programming languages, the algorithms underlying the drawing software of PIKAChU and SmilesDrawer are largely identical. We will briefly recap this algorithm below; more detailed descriptions of the algorithm’s elements can be found in the SmilesDrawer paper. [14]
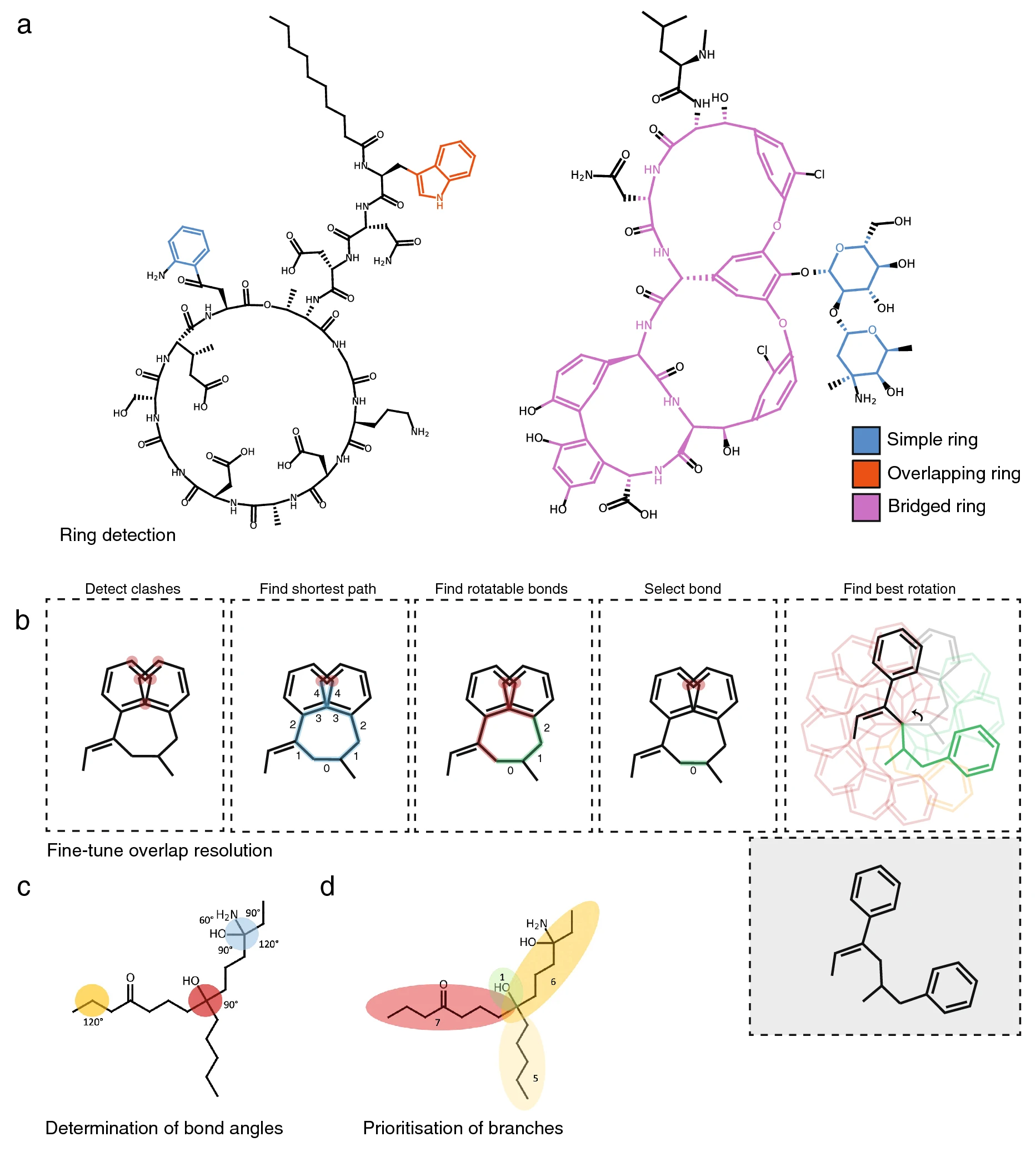
First, if indicated, PIKAChU’s drawing algorithm removes hydrogens from the graph. Next, it finds the smallest set of smallest rings in the structure graph. As SmilesDrawer’s SSSR implementation sometimes failed to detect some rings, leading to unreadable structure renderings (Additional file 2: Fig. S2), we implemented the SSSR algorithm ourselves. Next, like SmilesDrawer, PIKAChU classifies all rings into one of three groups: simple rings, overlapping rings, and bridged rings. Simple rings are standalone rings that do not have any overlapping atoms with any other rings. Overlapping rings are rings that overlap with one or more other rings, where the overlap between any two rings can comprise at most two atoms, any atom in the overlap is part of at most two rings, and no atoms in the ring overlap with bridged rings. Finally, bridged rings are rings that share more than two atoms with another ring, contain atoms that are part of three or more rings, or share atoms with another bridged ring (Fig. 2A).
|
After ring systems have been identified, atoms are placed onto a 2D coordinate system. If the molecule contains rings, positioning starts with the placement of an atom in a ring, prioritizing bridged rings over simple and overlapping rings. Then, the graph is traversed one atom at a time in depth-first fashion. If an atom is part of a ring, the entire ring or ring system get placed at once. In the case of simple and overlapping rings, ring placement can be done using simple polygon geometry. For bridged rings, atoms are positioned using the force-spring model described by Kamada and Kawai [9], where all atoms of the bridged system are initially placed in a circle, and then pulled towards their optimal positions by minimizing the difference between the desired bond length and the distance between neighboring atoms, and maximizing distances between non-neighboring atoms. Non-ring atoms are positioned a bond length away from the previous atom, where the angle with respect to the previous atom is determined by the number of neighbors the atom has (Fig. 2C), and the size of the molecular subtree behind each neighboring atom (Fig. 2D). Stereochemically restricted double bonds are always forced into the appropriate cis- or trans conformation. Unlike SmilesDrawer, which directly infers bond stereochemistry from the SMILES string, PIKAChU draws this information from bond objects stored in the molecular graph. As an improvement on SmilesDrawer, PIKAChU attempts to resolve wrongly depicted stereobonds in rings by mirroring one of the neighboring atoms into the ring. PIKAChU always selects the atom with the smallest protruding side chain for this purpose (Additional file 2: Fig. S3). When multiple consecutive stereobonds are found in a ring, PIKAChU adjusts them in order, never rotating a neighbor of the same bond twice.
Once all atoms have been assigned initial coordinates, atoms adjacent to rings are flipped outside of their ring where possible. Then, the drawing is checked for overlaps between atoms, and these overlaps are resolved by rotating branches of the molecule around single bonds. In PIKAChU, we have included an extra "fine-tuning" option that is not present in SmilesDrawer. When the fine-tuning flag is set to True, all pairs of clashing atoms are detected. Then, the shortest path is calculated between all clashing atoms. First, PIKAChU determines which bonds are rotatable: bonds are considered unrotatable when they are a chiral bond, are adjacent to a chiral bond, or are in a cycle. As rotations around bonds located equally far away from two clashing atoms likely have the greatest impact on clash resolution, PIKAChU selects the rotatable bond that is positioned as close to the center of the shortest path as possible. Next, PIKAChU takes the resulting set of bonds found for all clashes and rotates each at 30° intervals, assessing and storing the number of clashes in the drawing after each iteration. The angle for which the number of steric clashes is minimized is chosen (Fig. 2B).
Finally, some bonds adjacent to chiral centers are replaced with backward and forward wedges. They are placed such that they do not neighbor more than one chiral center where possible, they are not part of a ring, and point in the direction of the shortest branch leading from a chiral center, in that order of priority. The resulting image is subsequently written to an .svg or .png file or displayed directly in matplotlib.
Structure annotation
PIKAChU provides the option to add custom annotations to structures. Each Atom instance contains an "annotations" attribute, which points to an AtomAnnotation instance. An AtomAnnotations instance can contain as many annotations as the user requires. Annotations can be added to all atoms in a structure at once by defining the name of the attribute with a string and, optionally, providing a default value for the attribute. Subsequently, specific values can be added and retrieved for specific atoms or atom sets. A manual providing an example can be found on the PIKAChU wiki.
Substructure matching
PIKAChU detects occurrences of a substructure in a superstructure in five steps. In all steps, hydrogens are ignored. First, PIKAChU checks for each atom type in the substructure if enough atoms of these types are accounted for in the superstructure. Second, it assesses for each atom in the substructure whether an atom exists in the superstructure with the same connectivity, looking at directly neighboring bonds and atoms. Third, using the atom with the most diverse connectivity as a seed, it finds matches of the substructure in the superstructure using a depth-first search algorithm, ignoring stereochemistry. By first looking at atom type and atom connectivity, and by using atoms of diverse connectivity as seeds for substructure matching, the number of calls to the computationally expensive depth-first search function is minimized. Fourth, for each match, it determines if all chiral centres in the substructure have the same orientation as corresponding chiral centers in the superstructure. Fifth, PIKAChU checks if cis–trans orientation of double bonds in the substructure matches that of double bonds in the superstructure. Chiral center and double bond stereochemistry checks can be toggled by the user independently of one another. If chirality of bonds and atoms are considered, substructures with undefined stereochemistry will still match to parent structures with defined stereochemistry. This does not apply in reverse: if a stereocenter or stereobond is defined for a substructure, it will not match to parent structures with undefined stereochemistry.
The algorithm described is somewhat similar to the Ullmann algorithm [19], which first assesses if a candidate subgraph contains enough nodes of the correct degree prior to substructure matching and selects nodes of the most unique degree as seeds. A key difference is that PIKAChU’s substructure matching algorithm also takes into account the identity of a node’s neighbors, not just a node’s degree.
Substructures can be easily visualized through a range of functions in PIKAChU’s "general" 'library.
Fingerprinting
PIKAChU uses ECFP [15], an improved version of the classical Morgan fingerprinting that also takes into account cycle membership, to perform similarity searches and convert molecules to bit vectors for machine learning features. Using Python’s inbuilt hashlib library, PIKAChU initializes each atom to a 32-bit hash, derived from a tuple containing information on heavy neighbors, valence, atomic number, atomic weight, charge, hydrogen neighbors, and ring membership. Then, each atom hash is iteratively updated with hashes from its neighbors, as well as the distance from the neighbor to the atom and stereochemical information if the atom is a chiral center. The number of iterations depends on a radius which can be set to any number (default = 2 for ECFP-4 fingerprinting). (The ECFP algorithm was described in detail by Rogers and Hahn in 2010. [15]) Finally, duplicate hashes are removed, as well as different hashes representing the same substructure, yielding a set of 32-bit hashes that constitutes a molecule’s fingerprint.
Using ECFP fingerprinting, PIKAChU can calculate Jaccard/Tanimoto distance and/or similarity between any two molecules. Furthermore, PIKAChU can convert molecule libraries into bit vectors of varying lengths (default = 1024) and an accompanying list of substructures represented by those bit vectors that can be used in downstream machine learning algorithms.
Defining reaction targets
In order to facilitate implementation of reactions and reaction pathways, PIKAChU lets users define target bonds or atoms within substructures with a set of dedicated functions. These functions take a SMILES string representing a substructure, and either one or two integers that define an atom or a bond between two atoms, respectively. For example, the SMILES string "C(= O)NC," accompanied by the integers 0 (pointing to the first C atom) and 2 (pointing to the N atom), represents a peptide bond. The occurrences of these bonds/atoms are then detected within a superstructure through a substructure search and are returned as a list of bonds/atoms. Subsequently, the returned bonds/atoms can be used as reaction targets, for instance for bond hydrolysis or atom methylation, using functions in PIKAChU for breaking or creating bonds and adding or removing atoms. Reactions currently have to be encoded manually using a library of functions included in PIKAChU, which include functions for creating bonds, breaking bonds, adding and removing atoms, and splitting disconnected graphs into separate structures. We provided in-built condensation and hydrolysis functions, as well as a more elaborate ketoreductase function, as examples on our GitHub page.
Characterization and visualization of the polyketide ketoreduction reaction
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.