Journal:PIKAChU: A Python-based informatics kit for analyzing chemical units
| Full article title | PIKAChU: A Python-based informatics kit for analyzing chemical units |
|---|---|
| Journal | Journal of Cheminformatics |
| Author(s) | Terlouw, Barbara R.; Vromans, Sophie P.J.M.; Medema, Marnix H. |
| Author affiliation(s) | Wageningen University |
| Primary contact | barbara dot terlouw at wur dot nl |
| Year published | 2022 |
| Volume and issue | 14 |
| Page(s) | 34 |
| DOI | 10.1186/s13321-022-00616-5 |
| ISSN | 1758-2946 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://jcheminf.biomedcentral.com/articles/10.1186/s13321-022-00616-5 |
| Download | https://jcheminf.biomedcentral.com/track/pdf/10.1186/s13321-022-00616-5.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
As efforts to computationally describe and simulate the biochemical world become more commonplace, computer programs that are capable of in silico chemistry play an increasingly important role in biochemical research. While such programs exist, they are often dependency-heavy, difficult to navigate, or not written in Python, the programming language of choice for bioinformaticians.
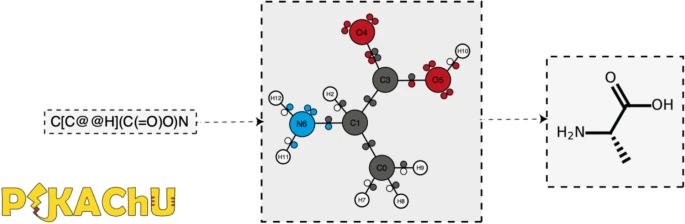
Here, we introduce PIKAChU (Python-based Informatics Kit for Analysing CHemical Units), a cheminformatics toolbox with few dependencies implemented in Python. PIKAChU builds comprehensive molecular graphs from SMILES strings, which allow for easy downstream analysis and visualization of molecules. While the molecular graphs (Graphical Abstract, below) PIKAChU generates are extensive—storing and inferring information on aromaticity, chirality, charge, hybridization, and electron orbitals—PIKAChU limits itself to applications that will be sufficient for most casual users and downstream Python-based tools and databases, such as Morgan fingerprinting, similarity scoring, substructure matching, and customizable visualization. In addition, it comes with a set of functions that assists in the easy implementation of reaction mechanisms. Its minimalistic design makes PIKAChU straightforward to use and install, in stark contrast to many existing toolkits, which are more difficult to navigate and come with a plethora of dependencies that may cause compatibility issues with downstream tools. As such, PIKAChU provides an alternative for researchers for whom basic cheminformatic processing suffices, and can be easily integrated into downstream bioinformatics and cheminformatics tools. (PIKAChU is available at https://github.com/BTheDragonMaster/pikachu.)
Keywords: cheminformatics kit, Python, structure visualization, in silico chemistry, molecular fingerprinting
Introduction
In a data-driven world where the discovery of novel natural and synthetic molecules is increasingly necessary, in silico chemical processing has become an essential part of biological and chemical research. Novel metabolites are compared or added to searchable chemical databases such as ChEBI [6], PubChem [10], NP Atlas [20], and COCONUT [17]; molecular structures are predicted from biological pathways [3, 16]; and bioactivities and pharmaceutical properties are predicted from chemical structure. [1, 18, 21] Such analyses rely on robust cheminformatics kits that can perform basic chemical processing, such as fingerprint-based similarity searches, substructure matching, molecule visualization, and chemical featurization for machine learning purposes.
Typically, molecular processing by cheminformatics kits begins with the reading in of molecular data from chemical data formats, ranging from one-dimensional to three-dimensional molecular representations. One such format is the SMILES (Simplified Molecular-Input Line Entry System) format, which represents a molecule as a one-dimensional string, describing atom composition, connectivity, stereochemistry, and charge. More elaborate formats such as PDB and MOL use text files to store not just the abovementioned properties but also atom coordinates in three-dimensional space.
Depending on the application, different formats and subsequent processing are appropriate. Due to the vast number of possible chemical analyses, exhaustive cheminformatics kits have accumulated into software libraries that are so large that they can be both hard to navigate and rely on so many dependencies that they can be difficult to implement in software packages. As a result, the trade-off between time spent accessing and integrating these cheminformatics kits into a codebase and time spent on actual analyses is disproportionate for users who need to perform simple in silico analyses such as reading in SMILES, drawing a molecule, or visualizing a substructure. One popular open-source cheminformatics kit that suffers from this problem is RDKit. [11] While RDKit is an incredibly fast and powerful library that supports an immense variety of possible chemical operations, its use of both Python and C++ as programming languages, as well as the sheer number of dependencies it relies on, frequently causes compatibility issues when integrating RDKit into other programs, and disproportionately increases the number of libraries that need to be installed. Therefore, while RDKit is great for heavy-duty in silico analyses such as computing 3D conformers for a compound or constructing electron density maps, it is a bit too much for the basic operations that most researchers in bioinformatics and cheminformatics require.
A second widely-used cheminformatics kit is CDK. [22] Written in Java, it is well-suited for implementation in web applications, and it has successfully been used for molecular processing in the COCONUT database [17], the Cytoscape application chemViz2 [13], and the scientific workflow platform KNIME (Konstanz Information Miner). [2] However, with Python becoming the programming language of choice for many scientists [4], especially those working in the growing field of (deep) neural networks, CDK is not always an ideal fit.
To make basic cheminformatics processing more accessible for Python programmers, we therefore introduce PIKAChU: a Python-based Informatics Kit for Analysing Chemical Units. PIKAChU is a flexible cheminformatics tool with few dependencies. It can parse molecules from SMILES, visualize chemical structures and substructures in matplotlib, perform extended connectivity fingerprinting (ECFP) [15] and Tanimoto similarity searches, and execute basic reactions with a focus on natural product chemistry. Therefore, we hope that PIKAChU can provide a convenient alternative for many Python-based bio- and cheminformatics tools and databases that only demand basic chemical processing.
Methods and implementations
Software description
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.