Journal:openBIS ELN-LIMS: An open-source database for academic laboratories
| Full article title | openBIS ELN-LIMS: An open-source database for academic laboratories |
|---|---|
| Journal | Bioinformatics |
| Author(s) | Barillari, Caterina; Ottoz, Diana S.M.; Fuentes-Serna, Juan M.; Ramakrishnan, Chandrasekhar; Rinn, Bernd; Rudolf, Fabian |
| Author affiliation(s) | ETH Zürich, Lifescience Zürich |
| Primary contact | Email: brinn at ethz dot ch -or- fabian dot rudolf at bsse dot ethz dot ch |
| Editors | Hancock, J. |
| Year published | 2015 |
| Volume and issue | 32 (4) |
| Page(s) | 638-640 |
| DOI | 10.1093/bioinformatics/btv606 |
| ISSN | 1460-2059 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://bioinformatics.oxfordjournals.org/content/32/4/638.long |
| Download | http://bioinformatics.oxfordjournals.org/content/32/4/638.full.pdf (PDF) |
Abstract
Summary: The open-source platform openBIS (open Biology Information System) offers an electronic laboratory notebook and a laboratory information management system (ELN-LIMS) solution suitable for the academic life science laboratories. openBIS ELN-LIMS allows researchers to efficiently document their work, to describe materials and methods and to collect raw and analyzed data. The system comes with a user-friendly web interface where data can be added, edited, browsed and searched.
Availability and implementation: The openBIS software, a user guide and a demo instance are available at https://openbis-eln-lims.ethz.ch. The demo instance contains some data from our laboratory as an example to demonstrate the possibilities of the ELN-LIMS.[1] For rapid local testing, a VirtualBox image of the ELN-LIMS is also available.
Introduction
Scientific recording is an essential part of research, as progress in science depends among other factors on existing knowledge and reproducibility. Recording is the process by which the scientific question, the choice of an experimental procedure, materials and methods, data analysis and interpretation of the results are gathered. Traditionally, all these details were kept in paper notebooks.
In today’s academic life science laboratories, most raw data, analyzed data and scripts are stored electronically, while the experimental details are often recorded in paper notebooks. This makes linking and retrieving experimental details and results problematic. Moreover, the large amount of data produced by today’s readout machines adds a level of complexity as the devices the data are stored on might soon be outdated.
Databases represent a solution to store all the necessary information together[2] because (i) they ensure long-lasting data storage by being independent of the operating environment; (ii) they are installed on a dedicated server, enabling long-term data storage and regular backups; (iii) they allow storage of large amounts of data; (iv) they are easily accessible via a local area network or the internet; (v) custom applications can be easily added.
Several commercial database solutions for ELNs are available, but they are not commonly adopted by academia. An internal evaluation among the research groups of the departments of Biosystems Science and Engineering and Biology of ETH Zurich found that the options affordable to academic labs had either unsuitable user interfaces (UI) or lack of features. The combination of an easy-to-use UI, flexible adaptation to a labs needs, support of an audit trail, physical support of the data server, support for managing large amounts of data and for integrating new measurement devices was only available at very high costs. On the other hand, the available open-source platforms lack many of the above features and, as they often originate from a single person within a research group, they also lack maintenance and support during the required data management life cycle.
Here, we describe the open-source database openBIS (open Biology Information System) electronic laboratory notebook and a laboratory information management system (ELN-LIMS), a software specifically developed with academic life science labs in mind. The structure of the database ensures that the results are represented in a logical and meaningful fashion, and it guarantees that the users can find, understand and reuse the data. openBIS ELN-LIMS consists of two interconnected parts: the LIMS, where the information about materials and methods is stored and kept up to date and the ELN, where data obtained from experiments are uploaded and annotated. The system is based on the openBIS platform[3], which, since 2007, is actively developed and maintained by a dedicated team of software engineers at the ETH and is productively used in several academic institutions and a few private companies.
Results
System's overview
Our goal was to implement a database to support the research activity of a typical biology laboratory. We conceived it as a tool to assist everyday activities: storage of materials, methods set-up, design and description of experiments, data labeling, storage and organization of results. To this purpose, we based our system on the platform openBIS[3], as it has the necessary features described above, is field-tested since about seven years and known to work well also with hundreds of terabytes of data.
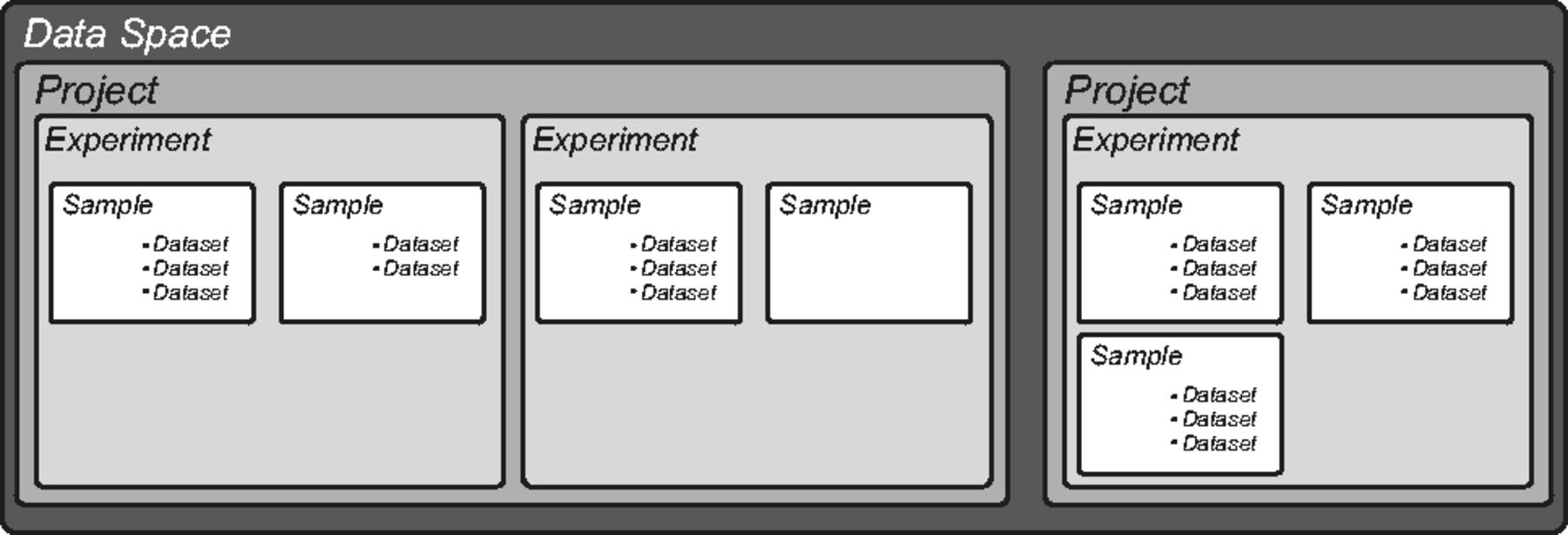
The openBIS core has a predefined hierarchical structure (Fig. 1). The top level, called "Data Space", controls access and rights and contains one or more "Projects," which in turn contain one or more "Experiments." "Samples" are the basic unit of openBIS and are organized within Experiments. We adapted this hierarchical structure to describe the organization and the objects of a typical life science laboratory. We defined the type and the number of Experiments and Samples and assigned customized properties to describe metadata. External files, in any kind of format, are uploaded as "Datasets" to each Sample. Each Sample can have any number of Datasets. We defined different types of Datasets and defined appropriate sets of properties (or annotations).
|
We used parent–child relationships linking Samples or Datasets to describe the logical or physical connections existing between the corresponding objects in the real world. We introduced the possibility to describe the nature of these relationships by annotating the links using texts or keywords.
To ensure that the creation, modification or deletion of any electronic records is traceable, computer-generated, time-stamped audit trails are used to record the date and time of the user’s intervention. In addition, Datasets are immutable, and as such all modifications made to the original data must be uploaded in separate Datasets using version numbers. Last, access to the system is restricted to authorized users, who can have different roles.[3] All these features are in accordance to the FDA regulations for electronic records.[4]
LIMS
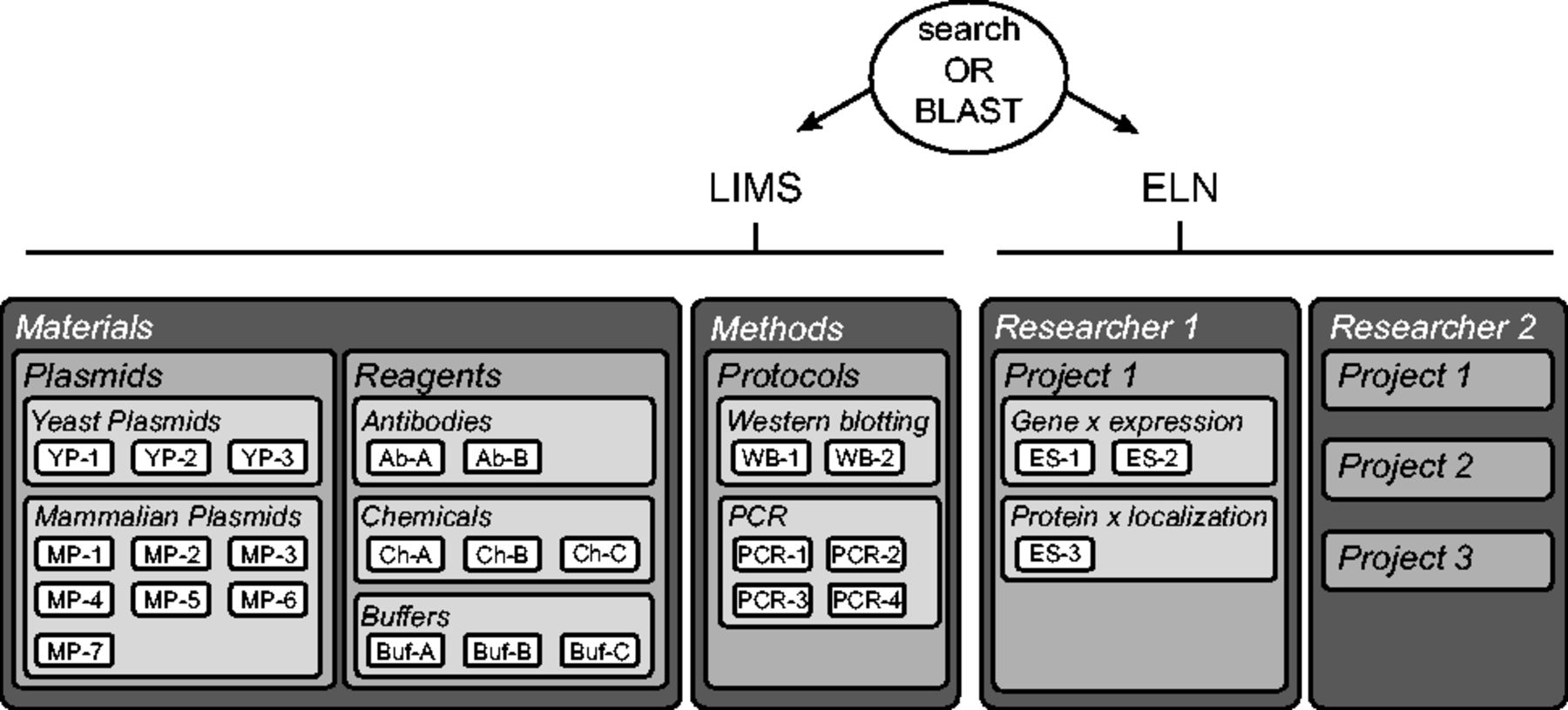
The LIMS consists of two "Data Spaces," the "Materials" and the "Methods," accessible and editable by all group members (Fig. 2 , left side).
|
The organization of the materials of each laboratory can be mapped to the structure of openBIS. Materials include plasmids, reagents, cell lines, etc. Each of them is a Sample type described by properties. For example, we have two plasmid collections: yeast and mammalian. In the Materials Data Space, we create a Project called "Plasmids" containing two Experiments: "Yeast Plasmids" and "Mammalian Plasmids." We upload Samples called Plasmids, having properties like plasmid name, backbone, marker, etc. We upload text files containing the DNA sequences to the corresponding plasmid entry as Datasets and integrate Plasmapper to visualize them as a plasmid map.[5]
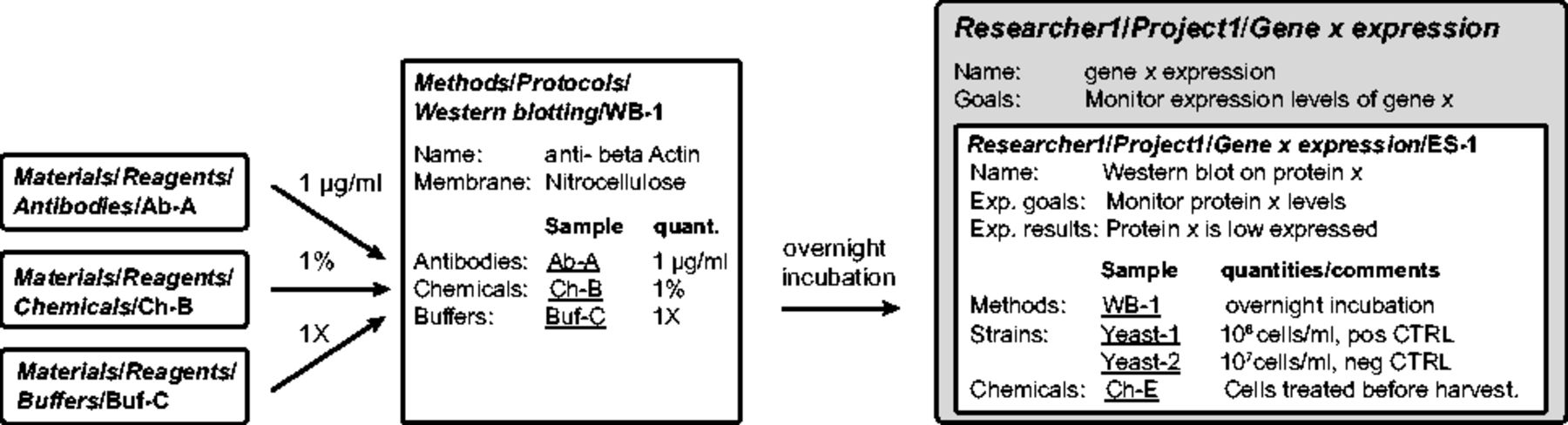
A method consists of a series of steps to accomplish a task, using several materials. Each method is described as a Sample type with specific properties. For example, we create a Sample type called "Western blotting" with properties like western blotting name, membrane, etc. We link western blotting Samples to material Samples, such as "Buffers," "Chemicals" and "Antibodies" and annotate the links with the quantities needed for the method (Fig. 3 ). For instance, we assign "anti-β-Actin" and "Nitrocellulose" to properties western blotting "Name" and "Membrane" in Western blotting 1. Then, we add 1 μg/ml of Antibody A and 1% of Chemical B in 1 × Buffer C. In the database, "Antibody A," "Chemical B" and "Buffer C" are added as parents of Western blotting 1 and are annotated with 1 μg/ml, 1% and 1×, respectively. Methods belonging to the same category are grouped in one Experiment. For example, we sort the western blotting and the PCR Samples into the western blotting Experiment and the PCR Experiment.
|
ELN
In the ELN, experimental data are stored and described in a scientifically meaningful manner, in terms of goal, method and interpretation of the results.
Each user has a personal Data Space where he/she can create and edit Projects, Experiments and Samples (Fig. 2, right side). An Experiment is a specific scientific question and contains Samples that are individual attempts to answer it. For example, to monitor the expression of gene x, we performed real-time quantitative PCR (RT-qPCR) and western blotting. In the database, we create an Experiment called "Gene × expression" and two Samples describing the results obtained by RT-qPCR and western blotting. All materials and methods used for each Sample are conveniently linked and can be annotated with specific details (e.g. deviation from the protocol). This ensures a complete description of the experiments and easy back-tracking (Fig. 3). The results of the RT-qPCR and the western blot can then be uploaded as Datasets to the corresponding Samples. Importantly, the information about configurations or set-up of the acquisition machines are stored in the Datasets as their settings are specific to this result. In our example, the raw data, the analyzed data and the analysis scripts are organized as separate Datasets.
Interaction with the system
An essential aspect of a database is its ease-of-use. We implemented an intuitive and easy user interface (ELN-UI) for the structure described above. This interface can be used on desktops and other devices, such as tablets. The ELN-UI main menu accesses the ELN, the LIMS, the Utilities (containing a storage manager), a generic search function and a BLAST search function.[6] Samples of the same type are listed in tables whose columns can be filtered with keywords. Single Samples can be selected to be visualized or printed separately. Connections between Samples can be inspected and browsed via a tree-like visualization.
We enter new Samples by filling in a form in the ELN-UI or via batch upload of text files. Data are uploaded as Datasets, either via the ELN-UI or via "dropbox scripts." These are written in Java or Jython and automatically register data and metadata, which can be directly imported from a data acquisition machine, like a microscope. Export of metadata and data is done with an export function from the ELN-UI or using dedicated scripts.
To set up the system, an administrator has to operate the previously described standard openBIS interface.[3] This interface is needed for configuration of the underlying database, as Data Spaces, Experiment, Sample and Dataset types and properties are created and maintained from there. However, normal users of the ELN-LIMS do not need to familiarize themselves with this interface as the ELN-UI possesses all necessary possibilities for everyday lab work and data retrieval.
Conclusion
We described an efficient ELN-LIMS based on the open source platform openBIS. This platform is pre-set with sensible defaults and ready for use. It can be customized and extended, as shown by the integration of PlasMapper and BLAST. Further extensions could be the integration of a function generating reports or the seamless integration of previously described openBIS extensions.[3]
Initial use of a database requires a time investment to organize the existing data, a learning phase and some security precautions. Nevertheless, we find that openBIS ELN-LIMS is of great help and worth the effort. Having all data stored in a single place and easily accessible sped up work and facilitated data retrieval in our laboratory.
Funding
The study was funded by the ETH Zürich.
Conflict of interest
None declared.
Additional notes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
Acknowledgements
We thank the yeast laboratory of the Stelling group for feedback during the implementation of the database and J. Stelling for his encouragement and support. We also thank F. van Drogen, C. Thoma, R. Denzler, M. Zimmermann, P. Markolin for their valuable feedback on the required functionality of the user interface, as well as testing of the implementation.
References
- ↑ Ottoz, D.S.; Rudolf, F.; Stelling, J. (2014). "Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae". Nucleic Acids Research 42 (17): e130. doi:10.1093/nar/gku616. PMC PMC4176152. PMID 25034689. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4176152.
- ↑ Bird, C.L.; Willoughby, C.; Frey, J.G. (2013). "Laboratory notebooks in the digital era: The role of ELNs in record keeping for chemistry and other sciences". Chemical Society Reviews 42 (20): 8157-75. doi:10.1039/c3cs60122f. PMID 23864106.
- ↑ 3.0 3.1 3.2 3.3 3.4 Bauch, A.; Adamczyk, I.; Buczek, P. et al. (2011). "openBIS: A flexible framework for managing and analyzing complex data in biology research". BMC Bioinformatics 12: 468. doi:10.1186/1471-2105-12-468. PMC PMC3275639. PMID 22151573. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3275639.
- ↑ "Electronic Code of Federal Regulations - Title 21: Food and Drugs - Part 11: Electronic Records; Electronic Signatures". U.S. Government Printing Office. http://www.ecfr.gov/cgi-bin/retrieveECFR?gp=&SID=04a3cb63d1d72ce40e56ee2e7513cca3&r=PART&n=21y1.0.1.1.8. Retrieved 25 October 2016.
- ↑ Dong, X.; Stothard, P.; Forsythe, I.J., Wishart, D.S. (2004). "PlasMapper: A web server for drawing and auto-annotating plasmid maps". Nucleic Acids Research 32 (Web Server issue): W660-4. doi:10.1093/nar/gkh410. PMC PMC441548. PMID 15215471. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC441548.
- ↑ Altschul, S.F.; Gish, W.; Miller, W. et al. (1990). "Basic local alignment search tool". Journal of Molecular Biology 215 (3): 403–10. doi:10.1016/S0022-2836(05)80360-2. PMID 2231712.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.