Journal:Terminology spectrum analysis of natural-language chemical documents: Term-like phrases retrieval routine
| Full article title | Terminology spectrum analysis of natural-language chemical documents: Term-like phrases retrieval routine |
|---|---|
| Journal | Journal of Cheminformatics |
| Author(s) | Alperin, Boris L.; Kuzmin, Andrey O.; Ilina, Ludmila Y.; Gusev, Vladimir D.; Salomatina, Natalia V.; Parmon, Valentin, N. |
| Author affiliation(s) | Boreskov Institute of Catalysis, Sobolev Institute of Mathematics, Novosibirsk State University |
| Primary contact | Email: kuzmin [at] catalysis.ru |
| Year published | 2016 |
| Volume and issue | 8 |
| Page(s) | 22 |
| DOI | 10.1186/s13321-016-0136-4 |
| ISSN | 1758-2946 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://jcheminf.springeropen.com/articles/10.1186/s13321-016-0136-4 |
| Download | http://jcheminf.springeropen.com/track/pdf/10.1186/s13321-016-0136-4 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
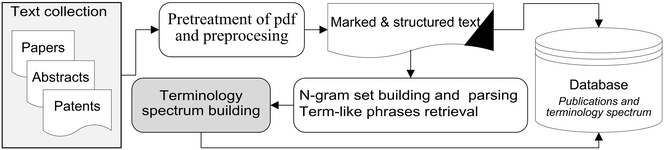
Background: This study seeks to develop, test and assess a methodology for automatic extraction of a complete set of ‘term-like phrases’ and to create a terminology spectrum from a collection of natural language PDF documents in the field of chemistry. The definition of ‘term-like phrases’ is one or more consecutive words and/or alphanumeric string combinations with unchanged spelling which convey specific scientific meanings. A terminology spectrum for a natural language document is an indexed list of tagged entities including: recognized general scientific concepts, terms linked to existing thesauri, names of chemical substances/reactions and term-like phrases. The retrieval routine is based on n-gram textual analysis with a sequential execution of various ‘accept and reject’ rules with taking into account the morphological and structural information.
Results: The assessment of the retrieval process, expressed quantitatively with a precision (P), recall (R) and F1-measure, which are calculated manually from a limited set of documents (the full set of text abstracts belonging to five EuropaCat events were processed) by professional chemical scientists, has proved the effectiveness of the developed approach. The term-like phrase parsing efficiency is quantified with precision (P = 0.53), recall (R = 0.71) and F1-measure (F1 = 0.61) values.
Conclusion: The paper suggests using such terminology spectra to perform various types of textual analysis across document collections. This sort of terminology spectrum may be successfully employed for text information retrieval, for reference database development, to analyze research trends in subject fields of research and to look for the similarity between documents.
|
|
Keywords: Terminology spectrum, natural language text analysis, n-Gram analysis, term-like phrases retrieval, text information retrieval
Background
The current situation in chemistry, as in any other field of natural science, can be characterized by a substantial growth of texts in natural languages (research papers, conference proceedings, patents, etc.), still being the most important sources of scientific knowledge and experimental data, information about modern research trends and terminology used in the subject areas of science. It greatly increases the value of such powerful information systems as Scopus®, SciFinder®, and Reaxys® which are capable of handling large text document databases and especially those fitted with advanced text information retrieval capabilities. In fact, both efficiency and productivity of modern scientific research in chemistry depend rigorously on quality and completeness of its information support, which is oriented firstly on advanced and flexible reference search, discovering and analysing of text information to afford the most relevant answers to user questions (substances, reactions, relevant patents or journal articles). The main ideas and developments in the information retrieval methods coupled with techniques of full text analysis are now well described and examined.[1]
In conventional information systems, the majority of text information retrieval and discovery methods are based on using specific sets of pre-defined document metadata, e.g. keywords or indexes of terms characterizing the texts content. User queries are converted using an index into information requests expressed by a combination of Boolean terms while bringing into play the vector space and terms weight. Probabilistic approaches may also be employed to take into account such features as terms distribution, co-occurrence information and their relationships derived from information retrieval thesauri (IRT) to include them into analytic process. Any kind of such indexes have to primarily be produced and updated manually by trained experts, but now the possibilities of automated index development attracts closer attention.
It is assumed that the structural foundation of any scientific text is its terminology, which may be represented, in principle, by advanced IRT. However, it leads to difficulties in applying conventional IRTs in practical information text analysis procedures because of limitations inherent in them. Typically, such thesauri are made manually in a very labor-intensive process and often are constructed to reflect the general terminology only. Terms from thesauri originally represent a formally written description of scientific conceptions and definitions which may not exactly match the real usage and spelling used in scientific texts. Moreover, a thesaurus developed for one type of text may be less efficient or not applicable when used with another. A good example is the IUPAC Gold Book compendium of chemical nomenclature, terminology, units and definition recommendations.[2] Terminology drafted by experts of IUPAC spans a wide range of chemistry but does not describe any field in detail and represents only a well-established upper level of scientific terminology. Summarizing, IRT based text analysis alone is unable to solve the problem of the variability of scientific texts written in natural languages because the accuracy of matching thesaurus terms with real text phrases leaves much to be desired.
It should also be noted that the language of science is evolving faster than that of natural language, especially in chemistry and molecular biology. Thus, the analysis of terminology of subject text collection should be done automatically using both primitive extraction and sophisticated knowledge-based parsing. Only automated data analysis can process and reveal the variety of term-like word combinations in the constantly changing world of scientific publications. Automated parsing and analysis of document collections or isolated documents for term-like phrases can also help to discover various contexts in which the same scientific terminology is used in different publications or even parts of the same publication.
There is nothing new in the idea of automated term retrieval. Typically, the terminology analysis of text content is focused on recognition of chemical entities and automatic keyphrase extraction aimed to provide a limited set of keywords which might characterize and classify the document as a whole. Two main strategies are usually applied: machine self-learning and usage of various dictionaries with automated selection rules (heuristics) coupled with calculated features[3], such as TF-IDF.[4][5] Therefore, keyphrase retrieval procedures typically involve the following stages: initial text pre-processing; selecting a candidate to a keyphrase; applying rules to each candidate; and compiling a list of keyphrases.[6] A few existing systems had been analyzed in terms of precision (P), recall (R) and F1-score attainable for existing keyphrase extraction datasets. For such well-known systems as Wingnus, Sztergak, and KP-Mminer, these values are reported as P = 0.34÷0.40, R = 0.11÷0.14, and F1 = 0.17÷0.20.[6] Open-Source Chemistry Analysis Routines (OSCAR4)[7] and ChemicalTagger[8] NLP may also be mentioned as tools for the recognition of named chemical entities and for parsing and tagging the language of text publications in chemistry.
However, there are some inherent shortcomings in the above mentioned keyphrase extraction approaches due to the presence of a significant amount of cases where a limited set of automatically selected top ranked keyphrases does not properly describe the document in details (e.g., a paper may contain the description of a specific procedure of catalyst preparation while not being the main subject of the paper). It may also be seen from the aforementioned values of P, R and F that in many cases the extracted keyphrases do not match the keyphrases selected by experts to an adequate degree. Exact matching of keyphrases is a rather rare event, partially due to the difficulties of taking into account nearly similar phrases, for instance, semantically similar phrases. On the other hand, even though the widely used n-gram analysis can build a full spectrum of token sequences present in the text, it may also produce a great level of noise, making it difficult to use them. Some attempts have been made to take into account the semantic similarity of n-grams and to differentiate between rubbish and candidates to plausible keyphrases.[9][10]
The problem of automatic recognition of scientific terms in natural language texts has been explored in recent decades.[11] That research has shown that taking into account the linguistic information may improve the terms extraction efficiency. The information about grammatical structure of multi-word scientific terms, their text variants, and the context of their usage may be represented as a set of lexico-syntactic patterns. For instance, values of P, R and F-measure equal to 73.1, 53.6 and 61.8 percent respectively for term extraction from scientific texts (only in Russian) on computer science and physics were obtained.[12]
References
- ↑ Salton, G. (1991). "Developments in Automatic Text Retrieval". pp. 974–980. doi:10.1126/science.253.5023.974. PMID 17775340.
- ↑ "IUPAC Gold Book". International Union of Pure and Applied Chemistry. 2014. http://goldbook.iupac.org/.
- ↑ Hussey, R.; Williams, S.; Mitchell, R. (2012). "Automatic keyphrase extraction: A comparison of methods". eKNOW, Proceedings of The Fourth International Conference on Information Process, and Knowledge Management: 18–23. ISBN 9781612081816.
- ↑ Eltyeb, S.; Salim, N. (2014). "Chemical named entities recognition: a review on approaches and applications". Journal of Cheminformatics 6: 17. doi:10.1186/1758-2946-6-17. PMC PMC4022577. PMID 24834132. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4022577.
- ↑ Gurulingappa, H.; Mudi, A.; Toldo, L.; Hofmann-Apitus, M.; Bhate, J. (2013). "Challenges in mining the literature for chemical information". RSC Advances 2013 (3): 16194-16211. doi:10.1039/C3RA40787J.
- ↑ 6.0 6.1 Kim, S.N.; Madelyan, O.; Kan, M.-Y.; Baldwin, T. (2013). "Automatic keyphrase extraction from scientific articles". Language Resources and Evaluation 47 (3): 723–742. doi:10.1007/s10579-012-9210-3.
- ↑ Jessop, D.M.; Adams, S.E.; Willighagen, E.L.; Hawizy, L.; Murray-Rust, P. (2011). "OSCAR4: A flexible architecture for chemical text-mining". Journal of Cheminformatics 3: 41. doi:10.1186/1758-2946-3-41. PMC PMC3205045. PMID 21999457. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3205045.
- ↑ Hawizy, L.; Jessop, D.M.; Adams, N.; Murray-Rust, P. (2011). "ChemicalTagger: A tool for semantic text-mining in chemistry". Journal of Cheminformatics 3: 17. doi:10.1186/1758-2946-3-17. PMC PMC3117806. PMID 21575201. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3117806.
- ↑ "Re-examining automatic keyphrase extraction approaches in scientific articles". MWE '09 Proceedings of the Workshop on Multiword Expressions: Identification, Interpretation, Disambiguation and Applications: 9–16. 2009. ISBN 9781932432602.
- ↑ "Approximate matching for evaluating keyphrase extraction". RANLP '09: International Conference on Recent Advances in Natural Language Processing: 484–489. 2009.
- ↑ Castellvi, M.T.C.; Bagot, R.E.; Palatresi, J.V. (2001). "Automatic term detection: A review of current systems". In Bourigault, D.; Jacquemin, C.; L'Homme, M.-C.. Recent Advances in Computational Terminology. John Benjamins Publishing Company. pp. 53–87. doi:10.1075/nlp.2.04cab. ISBN 9789027298164.
- ↑ Bolshakova, E.I.; Efremova, N.E. (2015). "A Heuristic Strategy for Extracting Terms from Scientific Texts". In Khachay, M.Y.; Konstantinova, N.; Panchenko, A.; Ignatov, D.I.; Labunets, V.G.. Analysis of Images, Social Networks and Texts. Springer International Publishing. pp. 297-307. doi:10.1007/978-3-319-26123-2_29. ISBN 9783319261232.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Numerous grammar errors were also corrected throughout the entire text.