File:Fig2 List ScientificReports2014 4.jpg
Original file (926 × 317 pixels, file size: 72 KB, MIME type: image/jpeg)
Summary
| Description |
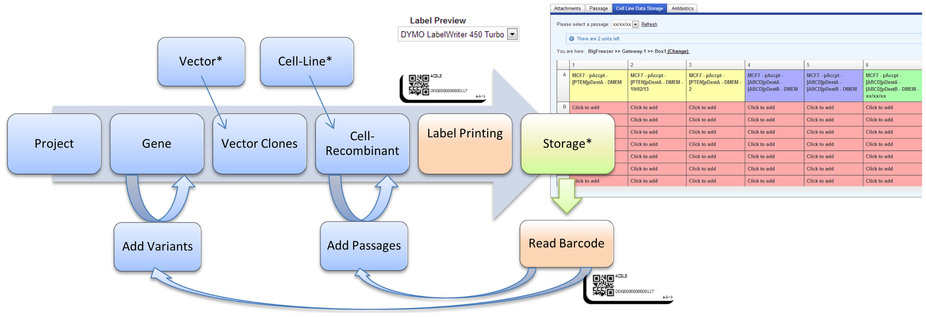
Figure 2. Initially, the administrators set up master data, such as vectors, cell-lines, medium compositions, as well as the storage infrastructure (*) Users then create projects and link genes to them. Vector clones are created from the genes, which in turn can be used to create cell-line recombinants. Samples are labeled using a DYMO label printer and added to physical, as well as virtual storage. At a later point, the barcode can be used for efficient retrieval and updating of sample information. Moreover, new gene variants and passages can be added with respective new labels and storage locations conveniently. Files and documents can be added to samples and genes, in order to make experimental results and additional information such as related publications, available to other users. |
|---|---|
| Source |
List, Markus; Schmidt, Steffen; Trojnar, Jakub; Thomas, Jochen; Thomassen, Mads; Kruse, Torben A.; Tan, Qihua; Baumbach, Jan; Mollenhauer, Jan (2014). "Efficient sample tracking with OpenLabFramework". Scientific Reports 4: 4278. doi:10.1038/srep04278. ISSN 2045-2322. http://www.nature.com/articles/srep04278. |
| Date |
2014 |
| Author |
Thomassen, Mads; Kruse, Torben A.; Tan, Qihua; Baumbach, Jan; Mollenhauer, Jan |
| Permission (Reusing this file) |
Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 20:03, 21 January 2016 | 926 × 317 (72 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









