File:Fig4 Calabria BMCBioinformatics2015 16-Suppl9.jpg
Original file (1,200 × 928 pixels, file size: 106 KB, MIME type: image/jpeg)
Summary
| Description |
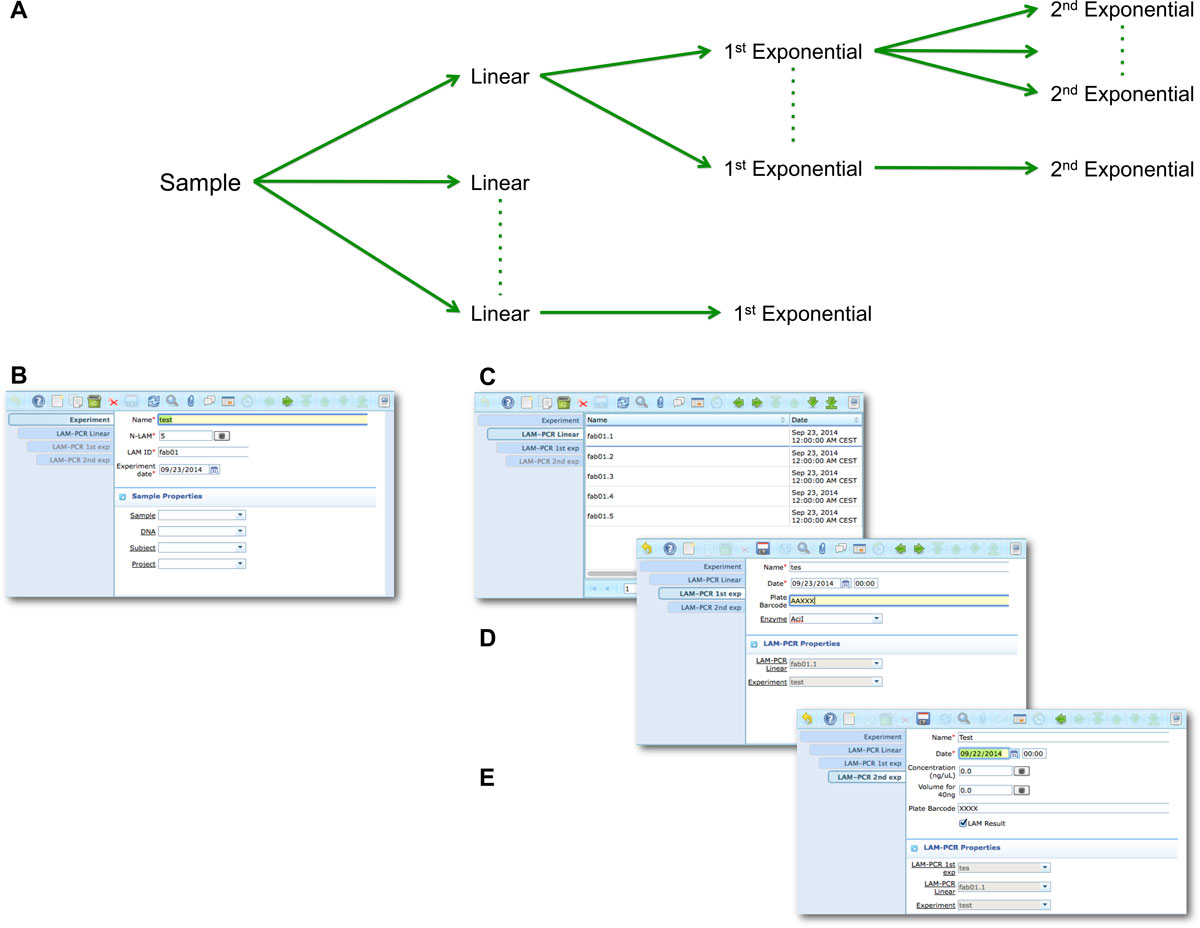
Figure 4: LAM-PCR workflow and the corresponding adLIMS automated process. The LAM-PCR procedure is reported as diagram (A) in which from a selected sample, different linear amplifications may be processed followed then by a first and a second exponential PCR amplification. The procedure is developed in adLIMS starting from the experiment set-up (B) in which a user fills the form with the data relative to the experiment such as "Name", number of LAM-PCRs (N-LAM), "LAM ID" and "date". Then adLIMS creates a number of entries as reported in the field "N-LAM", for the subsequent step "LAM-PCR linear" (C), that will be then processed and expanded in the "LAM-PCR 1st exp" step (D) with details such as "Name", "Plate barcode" and "Enzyme". The last step is the "LAM-PCR 2nd exp" (E), corresponding to the second exponential amplification, in which the user completes additional data of the experiment ("concentration" and "quality"). |
|---|---|
| Source |
Calabria, Andrea; Spinozzi, Giulio; Benedicenti, Fabrizio; Tenderini, Erika; Montini, Eugenio (2015). "adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies". BMC Bioinformatics 16 (Suppl 9): S5. doi:10.1186/1471-2105-16-S9-S5. ISSN 1471-2105. http://www.biomedcentral.com/1471-2105/16/S9/S5. |
| Date |
2015 |
| Author |
Calabria, Andrea; Spinozzi, Giulio; Benedicenti, Fabrizio; Tenderini, Erika; Montini, Eugenio |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 4.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 19:08, 16 December 2015 |  | 1,200 × 928 (106 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









