Journal:Shared metadata for data-centric materials science
| Full article title | Shared metadata for data-centric materials science |
|---|---|
| Journal | Scientific Data |
| Author(s) | Ghiringhelli, Luca M.; Baldauf, Carsten; Bereau, Tristan; Brockhauser, Sandor; Carbogno, Christian; Chamanara, Javad; Cozzini, Stefano; Curtarolo, Stefano; Draxl, Claudia; Dwaraknath, Shyam; Fekete, Ádám; Kermode, James; Koch, Christoph T.; Kühbach, Markus; Ladines, Alvin Noe; Lambrix, Patrick; Himmer, Maja-Olivia; Levchenko, Sergey V.; Oliveira, Micael; Michalchuk, Adam; Miller, Ronald E.; Onat, Berk; Pavone, Pasquale; Pizzi, Giovanni; Regler, Benjamin; Rignanese, Gian-Marco; Schaarschmidt, Jörg; Scheidgen, Markus; Schneidewind, Astrid; Sheveleva, Tatyana; Su, Chuanxun; Usvyat, Denis; Valsson, Omar; Wöll, Christof; Scheffler, Matthias |
| Author affiliation(s) | Friedrich-Alexander Universität, Humboldt-Universität zu Berlin, Fritz-Haber-Institut of the Max-Planck-Gesellschaft, University of Amsterdam, TIB – Leibniz Information Centre for Science and Technology and University Library, AREA Science Park, Duke University, Lawrence Berkeley National Laboratory, University of Warwick, Linköping University, Skolkovo Institute of Science and Technology, Max Planck Institute for the Structure and Dynamics of Matter, Federal Institute for Materials Research and Testing, University of Birmingham, Carleton University, École Polytechnique Fédérale de Lausanne, Paul Scherrer Institut, Chemin des Étoiles, Karlsruhe Institute of Technology, Forschungszentrum Jülich GmbH, University of Science and Technology of China, University of North Texas |
| Primary contact | Email: luca dot ghiringhelli at physik dot hu dash berlin dot de |
| Year published | 2023 |
| Volume and issue | 10 |
| Article # | 626 |
| DOI | 10.1038/s41597-023-02501-8 |
| ISSN | 2052-4463 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.nature.com/articles/s41597-023-02501-8 |
| Download | https://www.nature.com/articles/s41597-023-02501-8.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
The expansive production of data in materials science, as well as their widespread sharing and repurposing, requires educated support and stewardship. In order to ensure that this need helps rather than hinders scientific work, the implementation of the FAIR data principles (that ask for data and information to be findable, accessible, interoperable, and reusable) must not be too narrow. At the same time, the wider materials science community ought to agree on the strategies to tackle the challenges that are specific to its data, both from computations and experiments. In this paper, we present the result of the discussions held at the workshop on “Shared Metadata and Data Formats for Big-Data Driven Materials Science.” We start from an operative definition of metadata and the features that a FAIR-compliant metadata schema should have. We will mainly focus on computational materials science data and propose a constructive approach for the "FAIR-ification" of the (meta)data related to ground-state and excited-states calculations, potential energy sampling, and generalized workflows. Finally, challenges with the FAIR-ification of experimental (meta)data and materials science ontologies are presented, together with an outlook of how to meet them.
Keywords: materials science, data sharing, FAIR data principles, file formats, metadata, ontologies
Introduction: Metadata and FAIR data principles
The amount of data that has been produced in materials science up to today, and its day-by-day increase, are massive. [1] The dawn of the data-centric era [2] requires that such data are not just stored, but also carefully annotated in order to find, access, and possibly reuse them. Terms of good practice to be adopted by the scientific community for the management and stewardship of its data, the so-called FAIR data principles, have been compiled by the FORCE11 group. [3] Here, the acronym "FAIR" stands for "findable, accessible, interoperable, and reusable," which applies not only to data but also to metadata. Other terms for the “R” in FAIR are “repurposable” and “recyclable.” The former term indicates that data may be used for a different purpose than the original one for which they were created. The latter term hints at the fact that data in materials science are often exploited only once for supporting the thesis of a single publication, and then they are stored and forgotten. In this sense, they would constitute a “waste” that can be recycled, provided that they can be found and they are properly annotated.
Before examining the meaning and importance of the four terms of the FAIR acronym, it is worth defining what metadata are with respect to data. To that purpose, we start by introducing the concept of a data object, which represents the collective storage of information related to an elementary entry in a database. One can consider it as a row in a table, where the columns can be occupied by simple scalars, higher-order mathematical objects, strings of characters, or even full documents (or other media objects). In the materials science context, a data object is the collection of attributes (the columns in the above-mentioned table) that represent a material or, even more fundamentally, a snapshot of the material captured by a single configuration of atoms, or it may be a set of measurements from well-defined equivalent samples (see below for a discussion on this concept). For instance, in computational materials science, the attributes of a data object could be both the inputs (e.g., the coordinates and chemical species of the atoms constituting the material, the description of the physical model used for calculating its properties), and the outputs (e.g., total energy, forces, electronic density of states, etc.) of a calculation. Logically and physically, inputs and outputs are at different levels, in the sense that the former determine the latter. Hence, one can consider the inputs as metadata describing the data, i.e., the outputs. In turn, the set of coordinates A that are metadata to some observed quantities, may be considered as data that depend on another set of coordinates B, and the forces acting on the atoms in that set A. So, the set of coordinates B and the acting forces are metadata to the set A, now regarded as data. Metadata can always be considered to be data as they could be objects of different, independent analyses than those performed on the calculated properties. In this respect, whether an attribute of a data object is data or metadata depends on the context. This simple example also depicts a provenance relationship between the data and their metadata.
The above discussion can be summarized in a more general definition of the term metadata:
Metadata are attributes that are necessary to locate, fully characterize, and ultimately reproduce other attributes that are identified as data.
The metadata include a clear and unambiguous description of the data as well as their full provenance. This definition is reminiscent of the definition given by the National Institute of Standards and Technology (NIST) [4]: “Structured information that describes, explains, locates, or otherwise makes it easier to retrieve, use, or manage an information resource. Metadata is often called data about information or information about information.” With our definition, we highlight the role of data “reproducibility,” which is crucial in science.
Within the “full characterization” requirement, we highlight interpretation of the data as a crucial aspect. In other words, the metadata must provide enough information on a stored value (therein including, e.g., adimensional constants) to make it unambiguous whether two data objects may be compared with respect to the value of a given attribute or not.
Next, we should notice that, whereas in computational materials science the concept of data object identified by a single atomic configuration is well defined, in experimental materials science the concept of a class of equivalent samples is very hard to implement operationally. For instance, a single specimen can be altered by a measurement operation and thus cannot, strictly speaking, be measured twice. At the same time, two specimens prepared with the same synthesis recipe, may differ in substantial aspects due to the presence of different impurities or even crystal phases, thus yielding different values of a measured quantity. In this respect, here we use the term "equivalent sample" in its abstract, ideal meaning, but we also mention that one of the main purposes of introducing well-defined metadata in materials science is to provide enough characterization of experimental samples to put into practice the concept of equivalent samples.
The need for storing and characterizing data by means of metadata is determined by two main aspects, related to data usage. The first aspect is as old as science: reproducibility. In an experiment or computation, all the necessary information needed to reproduce the measured/calculated data (i.e., the metadata) should be recorded, stored, and retrievable. The second aspect becomes prominent with the demand for reusability. Data can and should be also usable for purposes that were not anticipated at the time they were recorded. A useful way of looking at metadata is that they are attributes of data objects answering the questions who, what, when, where, why, and how. For example, “Who has produced the data?”, “What are the data expected to represent (in physical terms)?”, “When were they produced?”, “Where are they stored?”, “For what purpose were they produced?”, and “By means of which methods were the data obtained?”. The latter two questions also refer to the concept of provenance, i.e., the logical sequence of operations that determine, ideally univocally, the data. Keeping track of the provenance requires the possibility to record the whole workflow that has lead to some calculated or measured properties (for more details, see the later section “Metadata for computational workflows”).
From a practical point of view, the metadata are organized in a schema. We summarize what the FAIR principles imply in terms of a metadata schema as follows:
- Findability is achieved by assigning unique and persistent identifiers (PIDs) to data and metadata, describing data with rich metadata, and registering (see below) the (meta)data in searchable resources. Widely known examples of PIDs are digital object identifiers (DOIs) and (permanent) Uniform Resource Identifiers (URIs). According to ISO/IEC 11179, a metadata registry (MDR) is a database of metadata that supports the functionality of registration. Registration accomplishes three main goals: identification, provenance, and monitoring quality. Furthermore, an MDR manages the semantics of the metadata, i.e., the relationships (connections) among them.
- Accessibility is enabled by application programming interfaces (APIs), which allow one to query and retrieve single entries as well as entire archives.
- Interoperability implies the use of formal, accessible, shared, and broadly applicable languages for knowledge representation (these are known as formal ontologies and will be discussed in the later section “Outlook on ontologies in materials science”), use of vocabularies to annotate data and metadata, and inclusion of references.
- Reusability hints at the fact that data in materials science are often exploited only once for a focus-oriented research project, and many data are not even properly stored as they turned out to be irrelevant for the focus. In this sense, many data constitute a “waste” that can be recycled, provided that the data can be found and they are properly annotated.
Establishing one or more metadata schemas that are FAIR-compliant, and that therefore enable the materials science community to efficiently share the heterogeneously and decentrally produced data, needs to be a community effort. The workshop “Shared Metadata and Data Formats for Big-Data Driven Materials Science: A NOMAD–FAIR-DI Workshop” was organized and held in Berlin in July 2019 to ignite this effort. In the following sections, we describe the identified challenges and first-stage plans, divided into different aspects that are crucial to be addressed in computational materials science.
In the next section, we describe the identified challenges and first plans for FAIR metadata schemas for computational materials science, where we also summarize as an example the main ideas behind the metadata schema implemented in the Novel-Materials Discovery (NOMAD) Laboratory for storing and managing millions of data objects produced by means of atomistic calculations (both ab initio and molecular mechanics), employing tens of different codes, which cover the overwhelming majority of what is actually used in terms of volume-of-data production in the community. We then follow with more detailed sections discussing the specific challenges related to interoperability and reusability for ground-state calculations (Section “Metadata for ground-state electronic-structure calculations”), perturbative and excited-state calculations (Section “Metadata for external-perturbation and excited-state electronic-structure calculations”), potential-energy sampling (molecular-dynamics and more, Section “Metadata for potential-energy sampling”), and generalized workflows (Section “Metadata for computational workflows”) are addressed in detail in the following sections. Challenges related to the choice of file formats are discussed in Section “File Formats.” An outlook on metadata schema(s) for experimental materials science and on the introduction of formal ontologies for materials science databases constitute Sections “Metadata schemas for experimental materials science” and “Outlook on ontologies in materials science,” respectively.
Towards FAIR metadata schemas for computational materials science
The materials science community has realized long ago that it is necessary to structure data by means of metadata schemas. In this section, we describe the pioneering and recent examples of such schemas, and how a metadata schema becomes FAIR-compliant.
To our knowledge, the first systematic effort to build a metadata schema for exchanging data in chemistry and materials science is CIF, an acronym that originally stood for "Crystallographic Information File," the data exchange standard file format introduced in 1991 by Hall, Allen and Brown. [5,6] Later, the CIF acronym was extended to also mean "Crystallographic Information Framework" [7], a broader system of exchange protocols based on data dictionaries and relational rules expressible in different machine-readable manifestations. These include the Crystallographic Information File itself, but also, for instance, XML (Extensible Markup Language), a general framework for encoding text documents in a format that is meant to be at the same time human and machine readable. CIF was developed by the International Union of Crystallography (IUCr) working party on Crystallographic Information and was adopted in 1990 as a standard file structure for the archiving and distribution of crystallographic information. It is now well established and is in regular use for reporting crystal structure determinations to Acta Crystallographica and other journals. More recently, CIF has been adapted to different areas of science such as structural biology (mmCIF, the macromolecular CIF [8]) and spectroscopy. [9] The CIF framework includes strict syntax definition in a machine-readable form and dictionary defining (meta)data items. It has been noted that the adoption of the CIF framework in IUCr publications has allowed for a significant reduction of the amount of errors in published crystal structures. [10,11]
An early example of an exhaustive metadata schema for chemistry and materials science is the Chemical Markup Language (CML) [12,13,14], whose first public version was released in 1995. CML is a dictionary, encoded in XML for chemical metadata. CML is accessible (for reading, writing, and validation) via the Java library JUMBO (Java Universal Molecular/Markup Browser for Objects). [14] The general idea of CML is to represent with a common language all kinds of documents that contain chemical data, even though currently the language—as of the latest update in 2012 [15]—covers mainly the description of molecules (e.g., IUPAC name, atomic coordinates, bond distances) and of inputs/outputs of computational chemistry codes such as Gaussian03 [16] and NWChem. [17] Specifically, in the CML representation of computational chemistry calculations [18], (ideally) all the information on a simulation that is contained in the input and output files is mapped onto a format that is in principle independent of the code itself. Such information is:
- Administrative data like the code version, libraries for the compilation, hardware, user submitting the job;
- Materials-specific (or materials-snapshot-specific) data like computed structure (e.g., atomic species, coordinates), the physical method (e.g., electronic exchange-correlation treatment, relativistic treatment), numerical settings (basis set, integration grids, etc.);
- Computed quantities (energies, forces, sequence of atomic positions in case a structure relaxation or some dynamical propagation of the system is performed, etc.).
The different types of information are hierarchically organized in modules, e.g., environment (for the code version, hardware, run date, etc.), initialization (for the exchange correlation treatment, spin, charge), molgeom (for the atomic coordinates and the localized basis set specification), and finalization (for the energies, forces, etc.). The most recent release of the CML schema contains more than 500 metadata-schema items, i.e., unique entries in the metadata schema. It is worth noticing that CIF is the dictionary of choice for the crystallography domain within CML.
Another long-standing activity is JCAMP-DX (Joint Committee on Atomic and Molecular Physical Data - Data Exchange) [19], a standard file format for exchange of infrared spectra and related chemical and physical information that was established in 1988 and then updated with IUPAC recommendations until 2004. It contains standard dictionaries for infrared spectroscopy, chemical structure, nuclear magnetic resonance (NMR) spectroscopy [20], and mass spectrometry [21], and ion-mobility spectrometry. [22] The European Theoretical Spectroscopy Facility (ETSF) File Format Specifications were proposed in 2007 [23,24,25], in the context of the European Network of Excellence NANOQUANTA, in order to overcome widely known portability issues of input/output file formats across platforms. The Electronic Structure Common Data Format (ESCDF) Specifications [26] is the ongoing continuation of the ETSF project and is part of the CECAM Electronic Structure Library, a community-maintained collection of software libraries and data standards for electronic-structure calculations. [27]
The largest databases of computational materials science data, AFLOW [28], Materials Cloud [29], Materials Project [30], the NOMAD Repository and Archive [31,32,33], OQMD [34], and TCOD [35] offer APIs that rely on dedicated metadata schemas. Similarly, AiiDA [36,37,38] and ASE [39], which are schedulers and workflow managers for computational materials science calculations, adopt their own metadata schema. OpenKIM [40] is a library of interatomic models (force fields) and simulation codes that test the predictions of these models, complemented with the necessary first-principles and experimental reference data. Within OpenKIM, a metadata schema is defined for the annotation of the models and reference data. Some of the metadata in all these schemas are straightforward to map onto each other (e.g., those related to the structure of the studied system, i.e., atomic coordinates and species, and simulation-cell specification), others can be mapped with some care. The OPTIMADE (Open Databases Integration for Materials Design [41]) consortium has recognized this potential and has recently released the first version of an API that allows users to access a common subset of metadata-schema items, independent of the schema adopted for any specific database/repository that is part of the consortium.
In order to clarify how a metadata schema can explicitly be FAIR-compliant, we describe as an example the main features of the NOMAD Metainfo, onto which the information contained in the input and output files of atomistic codes, both ab initio and force-field based, is mapped. The first released version of the NOMAD Metainfo is described by Ghiringhelli et al. [26] and it has powered the NOMAD Archive since the latter went online in 2014, thus predating the formal introduction of the FAIR data principles. [3]
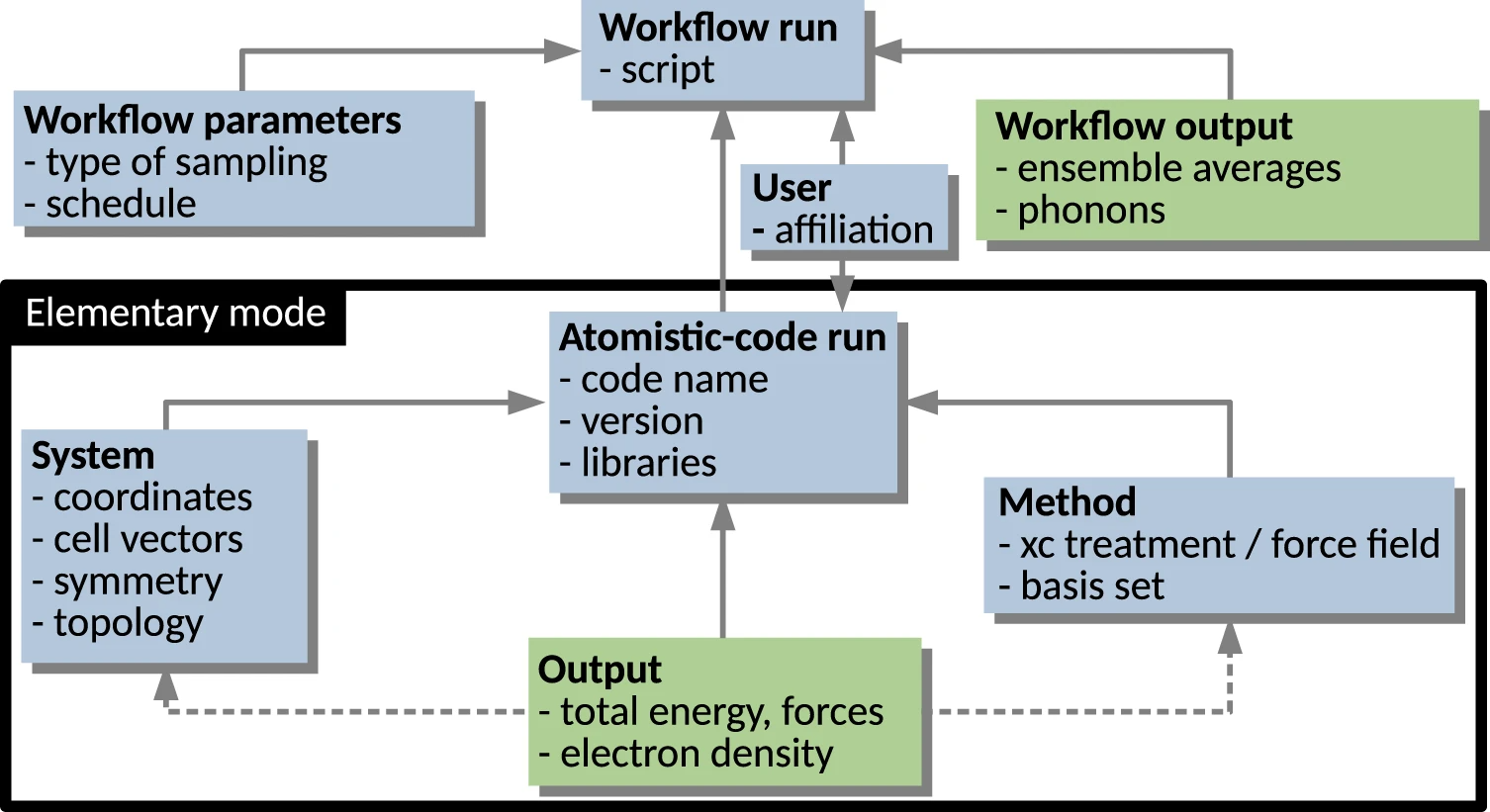
Here, we give a simplified description, graphically aided by Fig. 1, which highlights the hierarchical/modular architecture of the metadata schema. The elementary mode in which an atomistic materials science code is run (encompassed by the black rectangle) yields the computation of some observables (Output) for a given System, specified in terms of atomic species arranged by their coordinates in a box, and for a given physical model (Method), including specification of its numerical implementation. Sequences or collections of such runs are often defined via a Workflow. Examples of workflows are:
- Perturbative physical models (e.g., second-order Møller–Plesset, MP2, Green’s function based methods such as G0W0, random-phase approximation, RPA) evaluated using self-consistent solutions provided by other models (e.g., density-functional theory, DFT, Hartree-Fock method, HF) applied on the same System;
- Sampling of some desired thermodynamic ensemble by means of, e.g., molecular dynamics;
- Global- and local-minima structure searches;
- Numerical evaluations of equations of state, phonons, or elastic constants by evaluating energies, forces, and possibly other observables; and
- Scans over the compositional space for a given class of materials (high-throughput screening).
|
The workflows can also be nested, e.g., a scan over materials (different compositions and/or crystal structures) contains a local optimization for each material and extra calculations based on each local optimum structure such as evaluation of phonons, bulk modulus, or elastic constants, etc.
The NOMAD Metainfo organizes metadata into sections, which are represented in Fig. 1 by the labeled boxes. The sections are a type of metadata, which group other metadata, e.g., other sections or quantity-type metadata. The latter are metadata related to scalars, tensors, and strings, which represent the physical quantities resulting from calculations or measurements. In a relational database model, the sections would correspond to tables, where the data objects would be the rows, and the quantity-type metadata the columns. In its most simple realization, a metadata schema is a key-value dictionary, where the key is a name identifying a given metadata. In NOMAD Metainfo, similarly to CML, the key is a complex entity grouping the several attributes. Each item in NOMAD Metainfo has attributes, starting with its name, a string that must be globally unique, well defined, intuitive, and as short as possible. Other attributes are the human-understandable description, which clarifies the meaning of the metadata, the parent section, i.e., the section the metadata belongs to, and the type, whether the metadata is, e.g., a section or a quantity. Another possible type, the category type, will be discussed below. For the quantity-type metadata, other important attributes are physical units and shape, i.e., the dimensions (scalar, vector of a certain length, a matrix with a certain number of rows and columns, etc.), and allowed values, for metadata that admit only a discrete and finite set of values.
All definitions in the NOMAD Metainfo have the following attributes:
- A globally unique qualified name;
- Human-readable/interpretable description and expected format (e.g., scalar, string of a given length, array of given size);
- Allowed values;
- Provenance, which is realized in terms of a hierarchical and modular schema, where each data object is linked to all the metadata that concur to its definition. Related to provenance, an important aspect of NOMAD Metainfo is its extensibility. It stems from the recognition that reproducibility is an empirical concept, thus at any time, new, previously unknown or disregarded metadata may be recognized as necessary. The metadata schema must be ready to accommodate such extensions seamlessly.
The representation in Fig. 1 is very simplified for tutorial purposes. For instance, a workflow can be arbitrarily complex. In particular, it may contain a hierarchy of sub-workflows. In the currently released version of the NOMAD Metainfo, the elementary-code-run modality is fully supported, i.e., ideally all the information contained in a code run is mapped onto the metadata schema. However, the workflow modality is still under development. An important implication of the hierarchical schema is the mapping of any (complex) workflow onto the schema. That way, all the information obtained by its steps is stored. This is achieved by parsers, which have been written by the NOMAD team for each supported simulation code. One of the outcomes of the parsing is the assignment of a PID to each parsed data object, thus allowing for its localization, e.g., via a URI.
The NOMAD Metainfo is inspired by the CML, in particular in being hierarchical/modular. Each instance of a metadata-schema is uniquely identified, so that it can be associated with a URI for its convenient accessibility. An instance of a metadata schema can be generated by using a dedicated parser by pairing each parsed value with its corresponding metadata label. As an example, in Listing 1, we show a portion of the YAML file (see section “File Formats”) instantiating Metainfo for a specific entry of the NOMAD Archive. This entry can be searched by typing “entry_id = zvUhEDeW43JQjEHOdvmy8pRu-GEq” in the search bar at https://nomad-lab.eu/prod/v1/gui/search/entries. In Listing 1, key-value pairs are visible, as well as the nested-section structuring.
|
The modularity and uniqueness together allow for a straightforward extensibility, including customization, i.e., introduction of metadata-schema items that do not need to be shared among all users, but may be used by a smaller subset of users, without conflicts.
In Fig. 1, the solid arrows stand for the relationship is contained in between section-type metadata. A few examples of quantity-type metadata are listed in each box/section. Such metadata are also in an is-contained-in relationship with the section they are listed in. The dashed arrows symbolize the relationship has reference in. In practice, in the example of an Output section, the quantity-type metadata contained in such a section are evaluated for a given system described in a System section and for a given physical model described in a Method section. So, the section Output contains a reference to the specific System and Method sections holding the necessary input information. At the same time, the Output section is contained in a given Atomistic-code run section. These relationships among metadata already build a basic ontology, induced by the way computational data are produced in practice, by means of workflows and code runs. This aspect will be reexamined in the later Section “Outlook on ontologies in materials science.”
We now come to the category-type metadata that allow for complementary, arbitrarily complex ontologies to be built by starting from the same metadata. They define a concept, such as “energy” or “energy component,” in order to specify that a given quantity-type metadata has a certain meaning, be it physical (such as “energy”) or computer-hardware related, or administrative. To the purpose, each section and quantity-type metadata is related to a category-type metadata, by means of an is-a kind of relationship. Each category-type metadata can be related to another category-type metadata by means of the same is-a relationship, thus building another ontology on the metadata, which can be connected with top-down ontologies such as EMMO [42] (see Section “Outlook on ontologies in materials science” for a short description of EMMO).
The current version of NOMAD Metainfo includes more than 400 metadata-schema items. More specifically, these are the common metadata, i.e., those that are code-independent. Hundreds more metadata are code-specific, i.e., mapping pieces of information in the codes’ input/output that are specific to a given code and not transferable to other codes. The NOMAD Metainfo can be browsed at https://nomad-lab.eu/prod/v1/gui/analyze/metainfo.
To summarize, the NOMAD Metainfo addresses the FAIR data principles in the following sense:
- Findability is enabled by unique names and a human-understandable description;
- Accessibility is enabled by the PID assigned to each metadata-schema item, which can be accessed via a RESTful [43] API (i.e., an API supporting the access via web services, through common protocols, such as HTTP), specifically developed for the NOMAD Metainfo. Essentially all NOMAD data are open-access, and users who wish to search and download data do not need to identify themselves. They only need to accept the CC BY license. Uploaders can decide for an embargo. These data are then shared with a selected group of colleagues.
- Interoperability is enabled by the extensibility of the schema and the category-type metadata, which can be linked to existing and future ontologies (see Section “Outlook on ontologies in materials science”).
- Reusability/Repurposability/Recyclability is enabled by the modular/hierarchical structure that allows for accessing calculations at different abstraction scales, from the single observables in a code run to a whole complex workflow (see Section “Metadata for Computational Workflows”).
The usefulness and versatility of a metadata schema are demonstrated by the multiple access modalities it allows for. The NOMAD Metainfo schema is the basis of the whole NOMAD Laboratory infrastructure, which supports access to all the data in the NOMAD Archive, via the NOMAD API (also an implementation of the OPTIMADE API [41] is supported). This API powers three different access modes of the Archive: the Browser [44], which allows searches for single or groups of calculations, the Encyclopedia [45], which display the content of the Archive organized by materials, and the Artificial-Intelligence (AI) Toolkit [46,47,48], which connects in Jupyter Notebook's script-based queries and AI (machine learning, data mining) analyses of the filtered data. All the three services are accessible via a web browser running the dedicated GUI offered by NOMAD.
Metadata for ground-state electronic-structure calculations
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Several inline URLs from the original were turned into full citations for this version.