Journal:Effective information extraction framework for heterogeneous clinical reports using online machine learning and controlled vocabularies
| Full article title | Effective information extraction framework for heterogeneous clinical reports using online machine learning and controlled vocabularies |
|---|---|
| Journal | JMIR Medical Informatics |
| Author(s) | Zheng, Shuai; Lu, James. J.; Ghasemzadeh, Nima; Hayek, Salim S.; Quyyumi, Arshed, A.; Wang, Fusheng |
| Author affiliation(s) | Emory University, Stony Brook University |
| Primary contact | Email: fusheng dot wang at stonybrook dot edu; Phone: 1 6316327528 |
| Editors | Eysenbach, G. |
| Year published | 2017 |
| Volume and issue | 5 (2) |
| Page(s) | e12 |
| DOI | 10.2196/medinform.7235 |
| ISSN | 2291-9694 |
| Distribution license | Creative Commons Attribution 2.0 |
| Website | http://medinform.jmir.org/2017/2/e12/ |
| Download | http://medinform.jmir.org/2017/2/e12/pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: Extracting structured data from narrated medical reports is challenged by the complexity of heterogeneous structures and vocabularies and often requires significant manual effort. Traditional machine-based approaches lack the capability to take user feedback for improving the extraction algorithm in real time.
Objective: Our goal was to provide a generic information extraction framework that can support diverse clinical reports and enables a dynamic interaction between a human and a machine that produces highly accurate results.
Methods: A clinical information extraction system IDEAL-X has been built on top of online machine learning. It processes one document at a time, and user interactions are recorded as feedback to update the learning model in real time. The updated model is used to predict values for extraction in subsequent documents. Once prediction accuracy reaches a user-acceptable threshold, the remaining documents may be batch processed. A customizable controlled vocabulary may be used to support extraction.
Results: Three datasets were used for experiments based on report styles: 100 cardiac catheterization procedure reports, 100 coronary angiographic reports, and 100 integrated reports — each combines history and physical report, discharge summary, outpatient clinic notes, outpatient clinic letter, and inpatient discharge medication report. Data extraction was performed by three methods: online machine learning, controlled vocabularies, and a combination of these. The system delivers results with F1 scores greater than 95%.
Conclusions: IDEAL-X adopts a unique online machine learning–based approach combined with controlled vocabularies to support data extraction for clinical reports. The system can quickly learn and improve, thus it is highly adaptable.
Keywords: information extraction, natural language processing, controlled vocabulary, electronic medical records
Introduction
While immense efforts have been made to enable a structured data model for electronic medical records (EMRs), a large amount of medical data remain in free-form narrative text, and useful data from individual patients are usually distributed across multiple reports of heterogeneous structures and vocabularies. This poses major challenges to traditional information extraction systems, as either costly training datasets or manually crafted rules have to be prepared. These approaches also lack the capability of taking user feedback to adapt and improve the extraction algorithm in real time.
Our goal is to provide a generic information extraction framework that adapts to diverse clinical reports, enables a dynamic interaction between a human and a machine, and produces highly accurate results with minimal human effort. We have developed a system, Information and Data Extraction using Adaptive Online Learning (IDEAL-X), to support adaptive information extraction from diverse clinical reports with heterogeneous structures and vocabularies. The system is built on top of online machine learning and customizable controlled vocabularies. A demo video can be found on YouTube.[1]
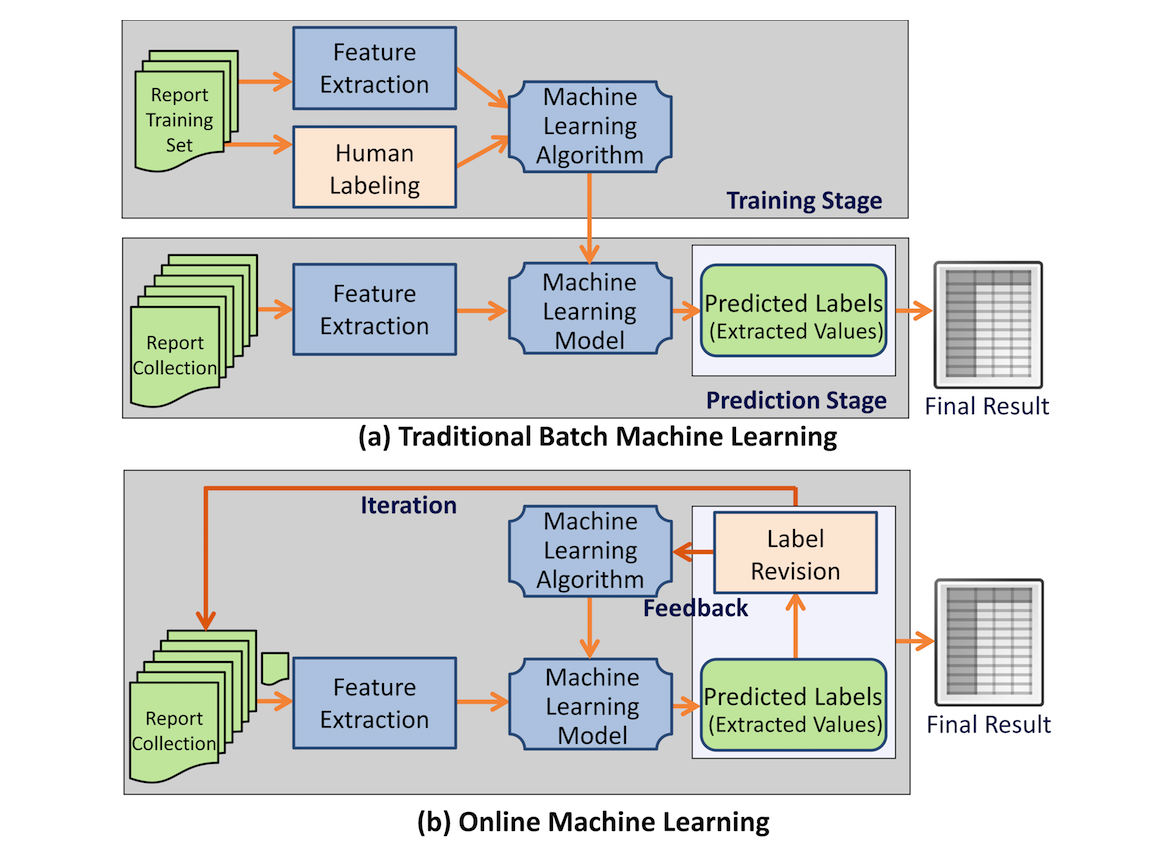
IDEAL-X uses an online machine learning–based approach[2][3][4] for information extraction. Traditional machine learning algorithms take a two-stage approach: batch training based on an annotated training dataset, and batch prediction for future datasets based on the model generated from stage one (Figure 1). In contrast, online machine learning algorithms[2][3] take an iterative approach (Figure 1). IDEAL-X learns one document at a time and predicts values to be extracted for the next one. Learning occurs from revisions made by the user, and the updated model is applied to prediction for subsequent documents. Once the model achieves a satisfactory accuracy, the remaining documents may be processed in batches. Online machine learning not only significantly reduces user effort for annotation but also provides the mechanism for collecting feedback from human-machine interaction to improve the system's model continuously.
Besides online machine learning, IDEAL-X allows for customizable controlled vocabularies to support data extraction from clinical reports, where a vocabulary enumerates the possible values that can be extracted for a given attribute. (The X in IDEAL-X represents the controlled vocabulary plug-in.) The use of online machine learning and controlled vocabularies is not mutually exclusive; they are complementary, which provides the user with a variety of modes for working with IDEAL-X.
|
Background
Related work
A number of research efforts have been made in different fields of medical information extraction. Successful systems include caTIES[5], MedEx [6], MedLEE[7], cTAKES[8], MetaMap[9], HITEx[10], and so on. These methods either take a rule-based approach, a traditional machine learning–based approach, or a combination of both.
References
- ↑ Zheng, S. (7 September 2014). "IDEAL-X: Information and Data Extraction using Adaptive Learning". YouTube. Google, Inc. https://www.youtube.com/watch?v=Q-DrWi31nv0. Retrieved 20 April 2017.
- ↑ 2.0 2.1 Shalev-Shwartz, S. (July 2007). "Online Learning: Theory, Algorithms, and Applications" (PDF). University of Chicago. http://ttic.uchicago.edu/~shai/papers/ShalevThesis07.pdf. Retrieved 03 May 2017.
- ↑ 3.0 3.1 Shalev-Shwartz, S. (2012). "Online Learning and Online Convex Optimization". Foundations and Trends in Machine Learning 4 (2): 107–194. doi:10.1561/2200000018.
- ↑ Smale, S.; Yao, Y. (2006). "Online Learning Algorithms". Foundations of Computational Mathematics 6 (2): 145–170. doi:10.1007/s10208-004-0160-z.
- ↑ Crowley, R.S.; Castine, M.; Mitchell, K. et al. (2010). "caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research". JAMIA 17 (3): 253-64. doi:10.1136/jamia.2009.002295. PMC PMC2995710. PMID 20442142. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2995710.
- ↑ Xu, H.; Stenner, S.P.; Doan, S. et al. (2010). "MedEx: A medication information extraction system for clinical narratives". JAMIA 17 (1): 19–24. doi:10.1197/jamia.M3378. PMC PMC2995636. PMID 20064797. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2995636.
- ↑ Friedman, C.; Shagina, L.; Lussier, Y.; Hripcsak, G. (2004). "Automated encoding of clinical documents based on natural language processing". JAMIA 11 (5): 392–402. doi:10.1197/jamia.M1552. PMC PMC516246. PMID 15187068. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC516246.
- ↑ Savova, G.K.; Masanz, J.J.; Ogren, P.V. et al. (2010). "Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications". JAMIA 17 (5): 507–13. doi:10.1136/jamia.2009.001560. PMC PMC2995668. PMID 20819853. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2995668.
- ↑ Aronson, A.R. (2001). "Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program". Proceedings AMIA Symposium 2001: 17–21. PMC PMC2243666. PMID 11825149. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2243666.
- ↑ Zeng, Q.T.; Goryachev, S.; Weiss, S. et al. (2006). "Extracting principal diagnosis, co-morbidity and smoking status for asthma research: Evaluation of a natural language processing system". BMC Medical Informatics and Decision Making 6: 30. doi:10.1186/1472-6947-6-30. PMC PMC1553439. PMID 16872495. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1553439.
Abbreviations
EMR: electronic medical record
HMM: hidden markov model
NLP: natural language processing
POS: part of speech
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In several cases the PubMed ID was missing and was added to make the reference more useful. Grammar and vocabulary were cleaned up to make the article easier to read.
Per the distribution agreement, the following copyright information is also being added:
©Shuai Zheng, James J Lu, Nima Ghasemzadeh, Salim S Hayek, Arshed A Quyyumi, Fusheng Wang. Originally published in JMIR Medical Informatics (http://medinform.jmir.org), 09.05.2017.