File:Fig3 Hatakeyama BMCBioinformatics2016 17.gif
Fig3_Hatakeyama_BMCBioinformatics2016_17.gif (778 × 307 pixels, file size: 57 KB, MIME type: image/gif)
Summary
| Description |
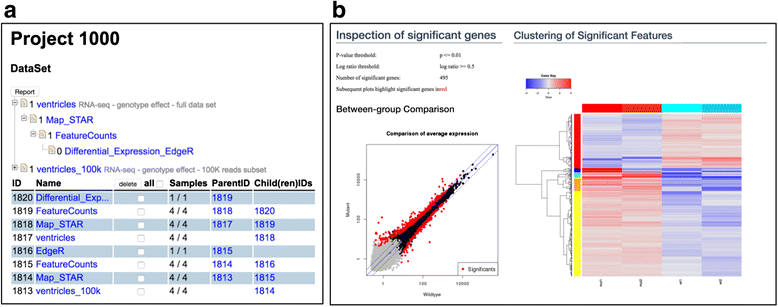
Figure 3. The screenshots of DataSet list and a part of a result generated by the edgeR SUSHI application. a. The DataSets are listed with a tree view (top) and table view (bottom). In the tree view, each node indicates a DataSet and the parental node indicates the input DataSet for the child node. b. Visualizations form the differential expression result the edgeR SUSHI application. We show a scatter plot with significantly differential expressed genes red-colored (left) and clustered heatmap (right). All calculated data is downloadable from this view. |
|---|---|
| Source |
Hatakeyama, M.; Opitz, L.; Russo, G.; Qi, W.; Schlapbach, R.; Rehrauer, H. (2016). "SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis". BMC Bioinformatics 17: 228. doi:10.1186/s12859-016-1104-8. |
| Date |
2016 |
| Author |
Hatakeyama, M.; Opitz, L.; Russo, G.; Qi, W.; Schlapbach, R.; Rehrauer, H. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 4.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 15:14, 6 July 2016 | 778 × 307 (57 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file: