Difference between revisions of "Journal:A proposed method of sample preparation and homogenization of hemp for the molecular analysis of cannabinoids"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Finished adding rest of content.) |
||
| Line 19: | Line 19: | ||
|download = [https://link.springer.com/content/pdf/10.1007/s42452-021-04747-6.pdf https://link.springer.com/content/pdf/10.1007/s42452-021-04747-6.pdf] (PDF) | |download = [https://link.springer.com/content/pdf/10.1007/s42452-021-04747-6.pdf https://link.springer.com/content/pdf/10.1007/s42452-021-04747-6.pdf] (PDF) | ||
}} | }} | ||
==Abstract== | ==Abstract== | ||
The use of ''[[Cannabis sativa]]'', or [[hemp]], in commercial, recreational, and pharmacological applications is on the rise in the United States and worldwide. Many of these applications have guidelines associated with them dependent on the concentration of [[cannabinoid]] molecules that keep the products classified as hemp versus marijuana, or that allow the producer to comment on the purity and [[potency]] of their product. Herein, we propose a method for homogenization of hemp that results in small particle sizes, uniform [[Sample (material)|samples]], and does not alter the cannabinoid concentrations during processing, allowing for optimal and reproducible potency testing. Using a novel “active grinding media,” we homogenized commercially available hemp to analyze approximately 100 mg samples of homogenate via sieve analysis and [[high-performance liquid chromatography]] (HPLC) to assess the resulting size and potency of the sample when using this methodology. When processing hemp samples with our proposed methodology, we have demonstrated the ability to produce 60.2% of all particles < 1.25 mm, with increased cannabinoid recovery compared to homogenates with larger average particle sizes. Maintaining sample temperatures below 35 °C during processing, we showed that our method does not thermally induce [[decarboxylation]] reactions that would result in major cannabinoid profile changes. We have developed a method for hemp processing via homogenization that does not alter the cannabinoid profile during processing, while consistently producing small particle sizes in a uniformly processed sample. This method allows for optimal and reproducible hemp processing when evaluating hemp and hemp-based products being brought to commercial markets. | The use of ''[[Cannabis sativa]]'', or [[hemp]], in commercial, recreational, and pharmacological applications is on the rise in the United States and worldwide. Many of these applications have guidelines associated with them dependent on the concentration of [[cannabinoid]] molecules that keep the products classified as hemp versus marijuana, or that allow the producer to comment on the purity and [[potency]] of their product. Herein, we propose a method for homogenization of hemp that results in small particle sizes, uniform [[Sample (material)|samples]], and does not alter the cannabinoid concentrations during processing, allowing for optimal and reproducible potency testing. Using a novel “active grinding media,” we homogenized commercially available hemp to analyze approximately 100 mg samples of homogenate via sieve analysis and [[high-performance liquid chromatography]] (HPLC) to assess the resulting size and potency of the sample when using this methodology. When processing hemp samples with our proposed methodology, we have demonstrated the ability to produce 60.2% of all particles < 1.25 mm, with increased cannabinoid recovery compared to homogenates with larger average particle sizes. Maintaining sample temperatures below 35 °C during processing, we showed that our method does not thermally induce [[decarboxylation]] reactions that would result in major cannabinoid profile changes. We have developed a method for hemp processing via homogenization that does not alter the cannabinoid profile during processing, while consistently producing small particle sizes in a uniformly processed sample. This method allows for optimal and reproducible hemp processing when evaluating hemp and hemp-based products being brought to commercial markets. | ||
| Line 53: | Line 48: | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="70%" | {| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="70%" | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px; | | colspan="6" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 1.''' For each listed set, three samples (''n'' = 3) were utilized at the listed weight, and the readings at each weight sieve were then averaged across all three samples to provide the values listed in this table. (''n'' = 3 STD ≤ ± 0.05) | ||
|- | |- | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" rowspan="2" | ! rowspan="2" style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Parameter | ||
! rowspan="2" style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Particle size distribution | |||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Weight sieve 2 | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Weight sieve 2 | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Weight sieve 7 | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Weight sieve 7 | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Weight sieve 12 | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Weight sieve 12 | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Weight sieve 17 | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Weight sieve 17 | ||
|- | |- | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|1.25 mm | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |1.25 mm | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|1.88 mm | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |1.88 mm | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|2.87 mm | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |2.87 mm | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|3.86 mm | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |3.86 mm | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px; | | rowspan="2" style="background-color:white; padding-left:10px; padding-right:10px;" |Set-1 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1.009 g (''n'' = 3) | | style="background-color:white; padding-left:10px; padding-right:10px;" |1.009 g (''n'' = 3) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.245 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.245 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.357 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.357 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.300 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.300 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.112 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.112 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Distribution (%) | | style="background-color:white; padding-left:10px; padding-right:10px;" |Distribution (%) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''24.4''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''24.4''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''35.5''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''35.5''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''29.0''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''29.0''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''11.2''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''11.2''' | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px; | | rowspan="2" style="background-color:white; padding-left:10px; padding-right:10px;" |Set-2 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1.006 g (''n'' = 3) | | style="background-color:white; padding-left:10px; padding-right:10px;" |1.006 g (''n'' = 3) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.490 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.490 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.414 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.414 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.069 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.069 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.035 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.035 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Distribution (%) | | style="background-color:white; padding-left:10px; padding-right:10px;" |Distribution (%) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''48.7''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''48.7''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''41.2''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''41.2''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''6.9''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''6.9''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''3.5''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''3.5''' | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px; | | rowspan="2" style="background-color:white; padding-left:10px; padding-right:10px;" |Set-3 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1.007 g (''n'' = 3) | | style="background-color:white; padding-left:10px; padding-right:10px;" |1.007 g (''n'' = 3) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.642 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.642 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.238 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.238 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.081 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.081 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.043 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.043 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Distribution (%) | | style="background-color:white; padding-left:10px; padding-right:10px;" |Distribution (%) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''63.85''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''63.85''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''23.69''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''23.69''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''8.08''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''8.08''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''4.24''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''4.24''' | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px; | | rowspan="2" style="background-color:white; padding-left:10px; padding-right:10px;" |Set-4 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1.007 g (''n'' = 3) | | style="background-color:white; padding-left:10px; padding-right:10px;" |1.007 g (''n'' = 3) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.606 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.606 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.268 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.268 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.106 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.106 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.025 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.025 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Distribution (%) | | style="background-color:white; padding-left:10px; padding-right:10px;" |Distribution (%) | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''60.239''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''60.239''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''26.673''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''26.673''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''10.537''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''10.537''' | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|'''2.485''' | | style="background-color:white; padding-left:10px; padding-right:10px;" |'''2.485''' | ||
|- | |- | ||
|} | |} | ||
| Line 135: | Line 130: | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="70%" | {| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="70%" | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px; | | colspan="9" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 2.''' Set-1, 5 m/s for 30 seconds. Set-2, 5 m/s for 60 seconds. Set-3, 5 m/s for 90 seconds. Set-4, 5 m/s for 120 seconds. For each listed set, three samples (''n'' = 3) were utilized at the listed weight and cannabinoid concentration of each were then averaged across all three samples to provide the values listed in this table. (''n'' = 3 STD ≤ ± 0.05) | ||
|- | |- | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px; | ! rowspan="2" style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Homogenization parameters | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px; | ! colspan="2" style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Cannabidiolic acid | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" colspan="2" | ! colspan="2" style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Cannabidiol | ||
! colspan="2" style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |∆<sup>9</sup>-Tetrahydrocannabinol | |||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px; | ! colspan="2" style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Tetrahydrocannabinolic acid | ||
|- | |- | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Average concentration (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Average concentration (%) | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Relative extraction efficiency (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Relative extraction efficiency (%) | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Average concentration (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Average concentration (%) | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Relative extraction efficiency (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Relative extraction efficiency (%) | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Average concentration (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Average concentration (%) | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Relative extraction efficiency (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Relative extraction efficiency (%) | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Average concentration (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Average concentration (%) | ||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;"|Relative extraction efficiency (%) | ! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Relative extraction efficiency (%) | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Set-1 | | style="background-color:white; padding-left:10px; padding-right:10px;" |Set-1 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|5.47 | | style="background-color:white; padding-left:10px; padding-right:10px;" |5.47 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.0 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1.92 | | style="background-color:white; padding-left:10px; padding-right:10px;" |1.92 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.0 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.16 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.16 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.0 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.15 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.15 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.0 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Set-2 | | style="background-color:white; padding-left:10px; padding-right:10px;" |Set-2 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|5.54 | | style="background-color:white; padding-left:10px; padding-right:10px;" |5.54 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1.3 | | style="background-color:white; padding-left:10px; padding-right:10px;" |1.3 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1.96 | | style="background-color:white; padding-left:10px; padding-right:10px;" |1.96 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2.1 | | style="background-color:white; padding-left:10px; padding-right:10px;" |2.1 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.16 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.16 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.0 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.15 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.15 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.0 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Set-3 | | style="background-color:white; padding-left:10px; padding-right:10px;" |Set-3 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|5.58 | | style="background-color:white; padding-left:10px; padding-right:10px;" |5.58 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |2.0 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2.22 | | style="background-color:white; padding-left:10px; padding-right:10px;" |2.22 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|15.6 | | style="background-color:white; padding-left:10px; padding-right:10px;" |15.6 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.16 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.16 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.0 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.17 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.17 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|13.3 | | style="background-color:white; padding-left:10px; padding-right:10px;" |13.3 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Set-4 | | style="background-color:white; padding-left:10px; padding-right:10px;" |Set-4 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|6.34 | | style="background-color:white; padding-left:10px; padding-right:10px;" |6.34 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|15.9 | | style="background-color:white; padding-left:10px; padding-right:10px;" |15.9 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2.36 | | style="background-color:white; padding-left:10px; padding-right:10px;" |2.36 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|22.9 | | style="background-color:white; padding-left:10px; padding-right:10px;" |22.9 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.20 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.20 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|25.0 | | style="background-color:white; padding-left:10px; padding-right:10px;" |25.0 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.16 | | style="background-color:white; padding-left:10px; padding-right:10px;" |0.16 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|6.7 | | style="background-color:white; padding-left:10px; padding-right:10px;" |6.7 | ||
|- | |- | ||
|} | |} | ||
|} | |} | ||
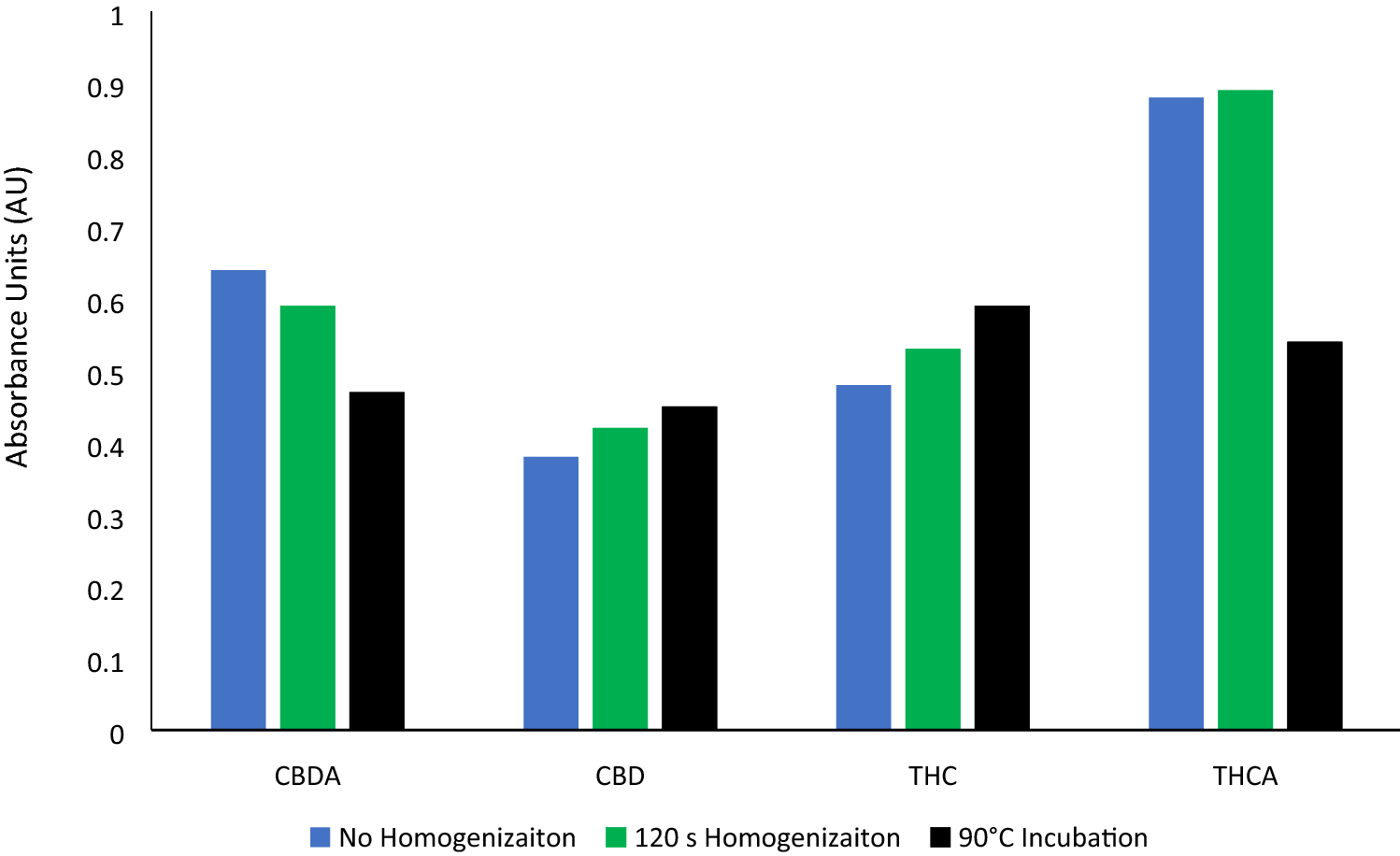
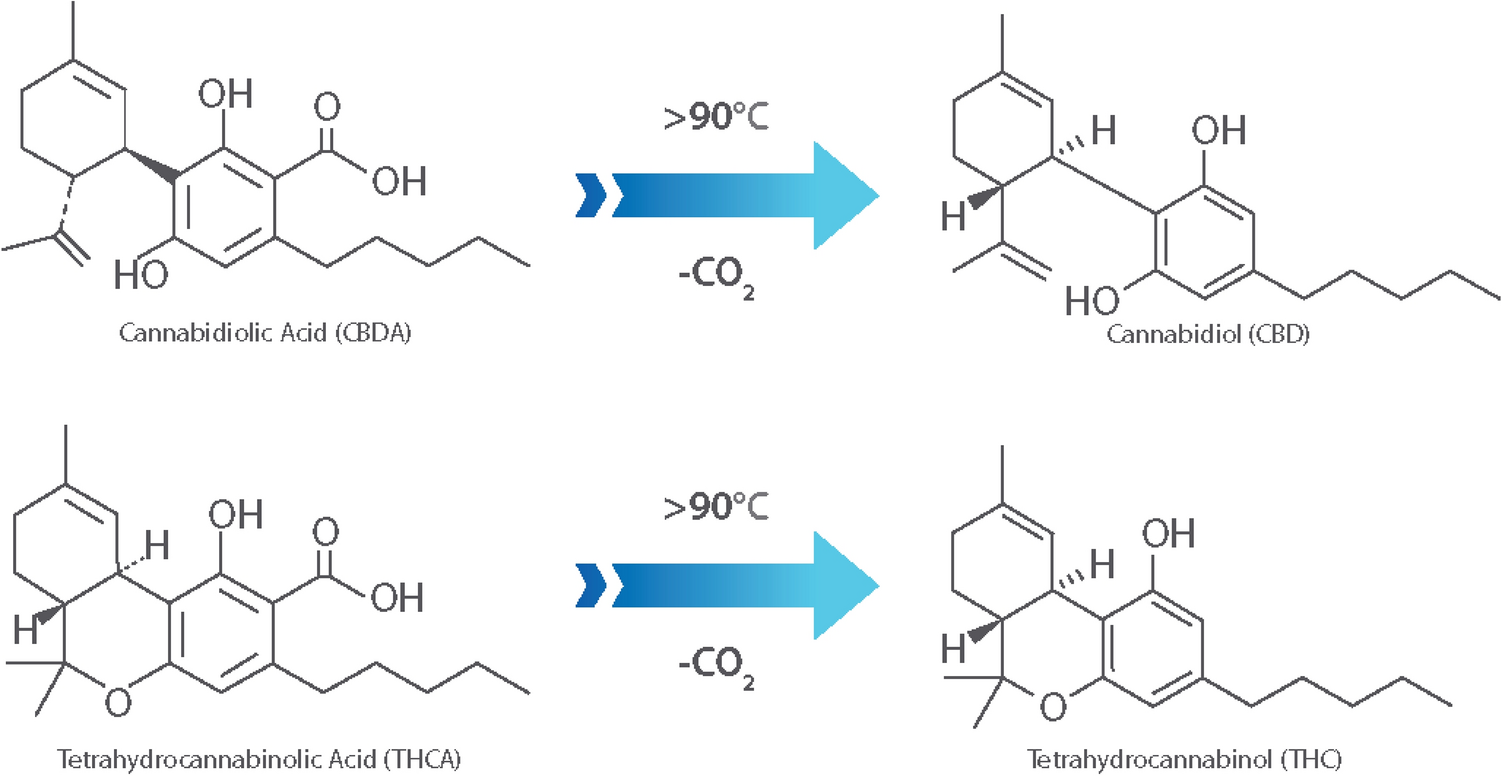
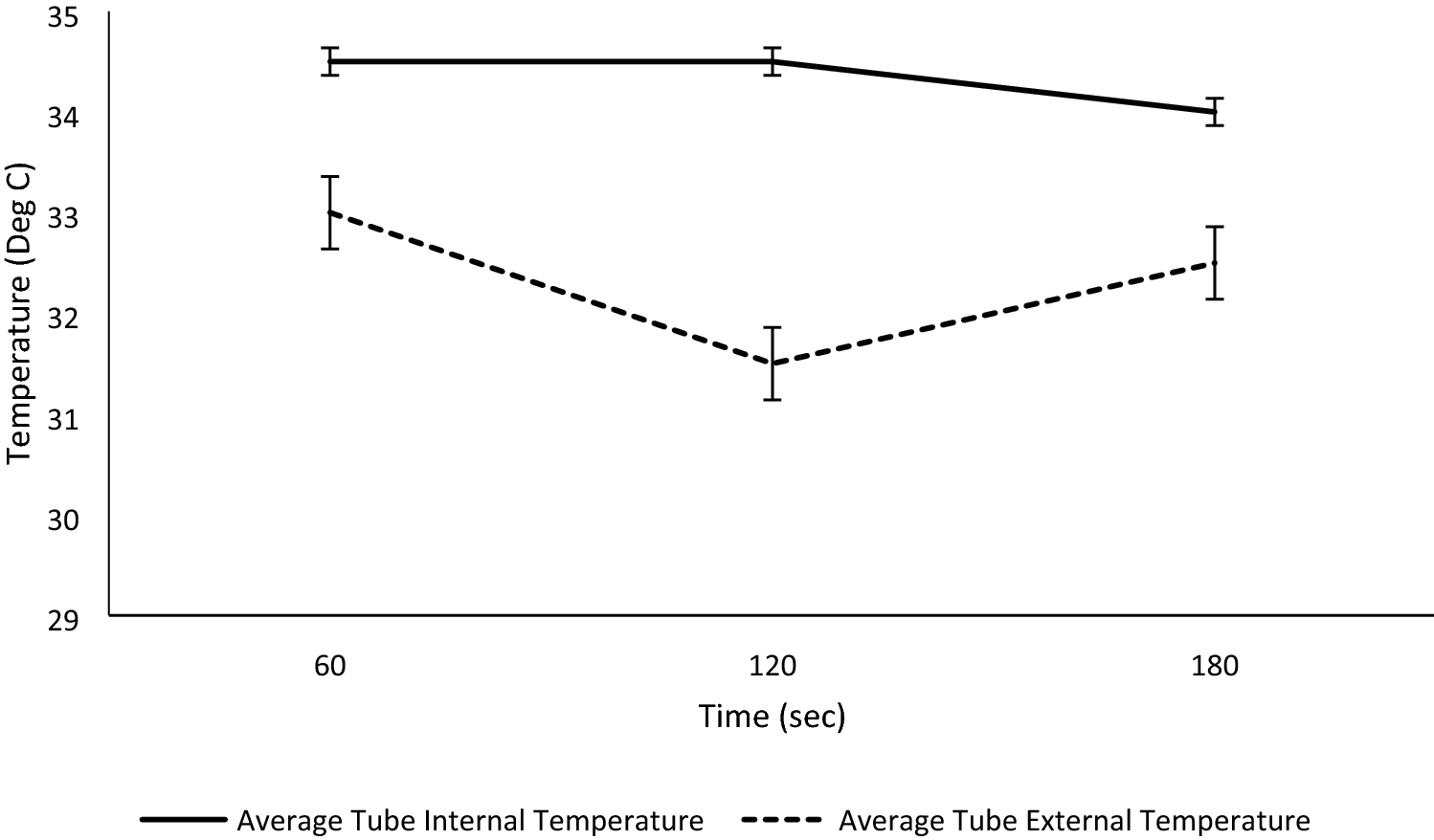
In addition to providing particle sizes < 1.25 mm, the use of our active grinding media and cyclical homogenization approach did not cause decarboxylation changes to the THCA and CBDA populations seen in the hemp tested, as confirmed by reverse phase HPLC (Fig. 1, | In addition to providing particle sizes < 1.25 mm, the use of our active grinding media and cyclical homogenization approach did not cause decarboxylation changes to the THCA and CBDA populations seen in the hemp tested, as confirmed by reverse phase HPLC (Fig. 1, Supplementary File 1). Processing hemp samples under these parameters did not produce enough thermal energy to alter the carboxylation start of the major cannabinoids tested for. Maintaining temperatures during sample processing under 90 °C is the benchmark to ensure that cannabinoids being tested for downstream do not have their concentrations altered during processing via decarboxylation reactions (Fig. 2). Additional homogenization runs with extended run times, up to 180 seconds, were also run in an attempt to determine the processing time required to generate enough heat to induce the decarboxylation reactions that convert THCA to THC and CBDA to CBD (Fig. 1, Supplemental Fig. 1). These extended time homogenizations showed that our method maintains processing temperatures under 35 °C, when measuring the temperature both internal and external to tubes used for the sample processing throughout the run (Fig. 3). | ||
| Line 207: | Line 202: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="700px" | {| border="0" cellpadding="5" cellspacing="0" width="700px" | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Fig. 1''' Reverse HPLC results visualizing CBDA, CBD, THCA, and THC concentrations as absorbance units following hemp homogenization (green), 90 C heating of sample (black), and no sample preparation (blue)</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Fig. 1''' Reverse HPLC results visualizing CBDA, CBD, THCA, and THC concentrations as absorbance units following hemp homogenization (green), 90 C heating of sample (black), and no sample preparation (blue)</blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 219: | Line 214: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="700px" | {| border="0" cellpadding="5" cellspacing="0" width="700px" | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Fig. 2''' CBD, CBDA, THC and THCA structure. Acid forms (CBDA and THCA) are decarboxylated to neutral forms when exposed to temperatures in excess of 90 °C, altering the cannabinoid profile of the processed hemp</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Fig. 2''' CBD, CBDA, THC and THCA structure. Acid forms (CBDA and THCA) are decarboxylated to neutral forms when exposed to temperatures in excess of 90 °C, altering the cannabinoid profile of the processed hemp</blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 231: | Line 226: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="700px" | {| border="0" cellpadding="5" cellspacing="0" width="700px" | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Fig. 3''' Temperature of homogenization tubes internally and externally was measured throughout the homogenization process. Note that the homogenization tubes never surpassed 35 C, maintaining the cannabinoids well under the 90 C benchmark that causes conversion from the acidic to neural forms.</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Fig. 3''' Temperature of homogenization tubes internally and externally was measured throughout the homogenization process. Note that the homogenization tubes never surpassed 35 C, maintaining the cannabinoids well under the 90 C benchmark that causes conversion from the acidic to neural forms.</blockquote> | ||
|- | |- | ||
|} | |} | ||
|} | |} | ||
==Discussion== | |||
The method proposed in this manuscript for hemp sample preparation via homogenization with our novel active grinding media demonstrates a critical step forward in the commercial and medical hemp markets. This methodology brings forward a standardized protocol for hemp processing that allows for reproducible results, small particle sizes, and unaltered cannabinoid profiles, all of which are essential as the market expands and inevitably becomes more standardized and regulated. While it is no surprise that federal, state, and local regulations have not developed as rapidly as the market for hemp-based products has, we must be fully aware that these regulations are coming and work proactively to establish robust industry standard methods for processing and testing hemp-involved products. | |||
The authors of this study firmly believe that this processing methodology is a strong first step towards establishing those industry standards for hemp processing and testing. Current guidelines surrounding hemp-derived products require that the measured THC levels remain below certain prescribed limits in order to remain labeled as hemp versus marijuana, a critical distinction in many markets across the United States where hemp is legal and marijuana is not.<ref name=":3" /><ref name=":5" /> While some argue that this is a game of legal semantics, this paradigm is critical for the producers of these products and compliance with these regulations is directly impacting their ability to remain in business. With such high stakes involved in accurate cannabinoid profiling, we were surprised to see so much variability in sample preparation techniques for these processes. The use of coffee grinders, blenders, and other various dissociation devices have two major problems seen when they are utilized for hemp sample preparation prior to cannabinoid profiling: 1. their dissociation processes are highly energetic, potentially heating the sample to a temperature to induce decarboxylation of THCA to THC, and 2. they are often very difficult to clean, allowing for cross contamination between samples.<ref>{{Cite journal |last=Atkins |first=Patricia L |date=2019-03-01 |title=Sample Processing and Preparation Considerations for Solid Cannabis Products |url=https://academic.oup.com/jaoac/article/102/2/427/5658162 |journal=Journal of AOAC INTERNATIONAL |language=en |volume=102 |issue=2 |pages=427–433 |doi=10.5740/jaoacint.18-0203 |issn=1060-3271}}</ref><ref>{{Cite journal |last=Pacifici |first=Roberta |last2=Marchei |first2=Emilia |last3=Salvatore |first3=Francesco |last4=Guandalini |first4=Luca |last5=Busardò |first5=Francesco Paolo |last6=Pichini |first6=Simona |date=2017-01-28 |title=Evaluation of cannabinoids concentration and stability in standardized preparations of cannabis tea and cannabis oil by ultra-high performance liquid chromatography tandem mass spectrometry |url=https://www.degruyter.com/document/doi/10.1515/cclm-2016-1060/html |journal=Clinical Chemistry and Laboratory Medicine (CCLM) |volume=55 |issue=10 |pages=1555–1563 |doi=10.1515/cclm-2016-1060 |issn=1437-4331}}</ref><ref>{{Cite journal |last=Ibrahim |first=Elsayed |last2=Wang |first2=Mei |last3=Radwan |first3=Mohamed |last4=Wanas |first4=Amira |last5=Majumdar |first5=Chandrani |last6=Avula |first6=Baharthi |last7=Wang |first7=Yan-Hong |last8=Khan |first8=Ikhlas |last9=Chandra |first9=Suman |last10=Lata |first10=Hemant |last11=Hadad |first11=Ghada |date=2019-03 |title=Analysis of Terpenes in Cannabis sativa L. Using GC/MS: Method Development, Validation, and Application |url=http://www.thieme-connect.de/DOI/DOI?10.1055/a-0828-8387 |journal=Planta Medica |language=en |volume=85 |issue=05 |pages=431–438 |doi=10.1055/a-0828-8387 |issn=0032-0943}}</ref> Both of these are ameliorated with our proposed methodology, implying a superior process for sample preparation, robust enough to be implemented into a standardized regulatory workflow. | |||
However, we do acknowledge that given the potential hundreds of coffee grinders and blenders in the current market which can be utilized for cannabis homogenization purposes, it would be impossible to compare each of them to our proposed methodology or even determine which is considered an “industry leader” brand in this market given the wide variability of techniques and lack of regulation. As such, we have elected to avoid direct comparisons with this proposed methodology. This purposeful decision of omission does not impact the utility of the proposed technology for hemp processing, but rather it further supports the need of a standardization of processing techniques to allow for future studies to benchmark against. | |||
In addition to the critical need for a standardized process that will not impact the THC profile of a hemp sample, it also important that the CBD profile be measured accurately.<ref name=":3" /><ref name=":5" /> The CBD profile of a hemp-based product is often tied to the potency of the product, with higher concentrations of CBD fetching higher prices in the market. Ensuring that accurate measurement of these cannabinoids is done requires a process that does not induce excessive thermal energy to the sample and produces a reliably small particle size throughout the sample to ensure accurate measurement with HPLC. | |||
As the hemp market continues to exponentially grow across the U.S., we feel that it is a pertinent time to adopt industry-standardized procedures. The significance of this work is to show that homogenization is a dependable and consistent method for measuring cannabinoid potency in hemp. With many states legally allowing hemp (''Cannabis'' with less than 0.3% THC) usage but prohibiting the use of cannabis with high levels of THC, it is critical to develop processing technologies that will not alter these cannabinoid profiles. The conversion of THCA to active THC can redefine a hemp product, into a high THC cannabis product, rendering it illegal. Having an accurate measurement of all forms of cannabinoids is important to prevent this. We show that the smaller the particles of hemp the more accurate potency testing becomes. Furthermore, this method is repeatable, and no contamination from multiple samples occurs. Having accurate potency testing also helps growers market their premium product verses product of lesser grades of potency. The method described in this manuscript is a robust approach to hemp sample preparation which allows for accurate and reproducible downstream molecular analysis of each sample’s cannabinoid profile. | |||
==Conclusions== | |||
Herein, we are proposing a methodology for hemp sample preparation and homogenization that provides uniform homogenization of a sample, while maintaining the integrity and proportionality of the cannabinoids for downstream molecular analysis. Through use of the active grinding media and bead-mill homogenization, we prepared samples with a majority of the particles < 1.25 mm, resulting in increased cannabinoid recovery when compared to larger particle size homogenates, and no alterations in the carboxylation profiles of CBDA or THCA during processing. This method provides critical reproducibility to processing hemp, allowing the end user to maintain the highest of potency and purity standards when moving forward with their molecular analysis. | |||
==Supplementary information== | |||
* [https://static-content.springer.com/esm/art%3A10.1007%2Fs42452-021-04747-6/MediaObjects/42452_2021_4747_MOESM1_ESM.docx Supplementary File 1] (.docx) | |||
==Abbreviations== | |||
* THCA: Δ<sup>9</sup>-Tetrahydrocannabinolic acid | |||
* THC: Tetrahydrocannabinol | |||
* CBDA: Cannabidiolic acid | |||
* CBD: Cannabidiol | |||
* HPLC: High-performance liquid chromatography | |||
==Acknowledgements== | |||
The authors would like to acknowledge Karl Jahn, Pete Tortorelli, and Erik Masefield of Omni International, Inc. for their financial commitment and continued support of our research. Additionally, we would like to acknowledge Rachele Capobianco and Derick Gordon for their support in establishing the collaborations needed for this project. Finally, we would like to acknowledge Rachel True, Rachel Nash, and Leah Proctor for their sacrifices and unwavering support of our research. IANAFS. | |||
===Contributions=== | |||
ZPM: formal analysis, writing—original draft, review and editing, visualization. GLR: investigation, formal analysis, writing—review and editing. CMP: methodology, investigation, writing—review and editing. AO: investigation. WT: methodology, investigation. DB: methodology, writing—review and editing, supervision, project administration. TW: investigation. SP: resources, supervision, funding acquisition. BM: investigation. VC investigation. BE: conceptualization, methodology, investigation, data curation, writing—reviewing and editing. JA: conceptualization, supervision, funding acquisition. RJN: conceptualization, investigation, resources, writing—original draft, review and editing, visualization, supervision, project administration, funding acquisition. YCP: conceptualization, investigation, resources, data curation, visualization, supervision, project administration. | |||
===Funding=== | |||
This project was funded in its entirety by the private entities Omni International, Inc. and AMERICANNA Laboratories, LLC. | |||
===Availability of data and materials=== | |||
All data used in the development of this manuscript is available through email requests to authors Z.P. Morehouse or R.J. Nash at zmorehouse at omni-inc dot com or rnash at omni-inc dot com respectively. | |||
===Conflict of interest=== | |||
G.L. Ryan, C.M. Proctor, A. Okparanta, W. Todd, D. Bunting, T. White, B. Easparro, and J. Atwood are all employed by Omni International, Inc. but do not have any personal financial interests in the success of the company. Z.P. Morehouse has a consultancy contract with Omni International, Inc. but does not have any personal financial interests in the success of the company. R.J. Nash has a consultancy contract with Omni International, Inc. but does not have any personal financial interests in the success of the company, and is co-founder and owner of Jeevan Biosciences (Tucker, GA USA) to which he does have financial interests in its success. B. Miller, V. Colon, and Y.C. Park are all employed by AMERICANNA Laboratories, LLC and do not have any personal financial interests in its success. S. Perez is cofounder and co-owner of AMERICANNA Laboratories, LLC to which he does have personal financial interests in its success. | |||
==References== | ==References== | ||
Revision as of 22:51, 25 August 2021
| Full article title | A proposed method of sample preparation and homogenization of hemp for the molecular analysis of cannabinoids |
|---|---|
| Journal | SN Applied Sciences |
| Author(s) | Morehouse, Zachary P.; Ryan, Gabriella L.; Proctor, Caleb M.; Okparanta, Akelachi; Todd, Will; Bunting, Derrick; White, Taylor; Parez, Steven; Miller, Blessida; Colon, Veronica; Easparro, Brandon; Atwood, James; Park, YoungChul; Nash, Rodney J. |
| Author affiliation(s) | Omni International, Jeevan Biosciences, Georgia State University, AMERICANNA Laboratories |
| Primary contact | Email: Online form |
| Year published | 2021 |
| Volume and issue | 3 |
| Article # | 762 |
| DOI | 10.1007/s42452-021-04747-6 |
| ISSN | 2523-3971 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://link.springer.com/article/10.1007%2Fs42452-021-04747-6 |
| Download | https://link.springer.com/content/pdf/10.1007/s42452-021-04747-6.pdf (PDF) |
Abstract
The use of Cannabis sativa, or hemp, in commercial, recreational, and pharmacological applications is on the rise in the United States and worldwide. Many of these applications have guidelines associated with them dependent on the concentration of cannabinoid molecules that keep the products classified as hemp versus marijuana, or that allow the producer to comment on the purity and potency of their product. Herein, we propose a method for homogenization of hemp that results in small particle sizes, uniform samples, and does not alter the cannabinoid concentrations during processing, allowing for optimal and reproducible potency testing. Using a novel “active grinding media,” we homogenized commercially available hemp to analyze approximately 100 mg samples of homogenate via sieve analysis and high-performance liquid chromatography (HPLC) to assess the resulting size and potency of the sample when using this methodology. When processing hemp samples with our proposed methodology, we have demonstrated the ability to produce 60.2% of all particles < 1.25 mm, with increased cannabinoid recovery compared to homogenates with larger average particle sizes. Maintaining sample temperatures below 35 °C during processing, we showed that our method does not thermally induce decarboxylation reactions that would result in major cannabinoid profile changes. We have developed a method for hemp processing via homogenization that does not alter the cannabinoid profile during processing, while consistently producing small particle sizes in a uniformly processed sample. This method allows for optimal and reproducible hemp processing when evaluating hemp and hemp-based products being brought to commercial markets.
Keywords: hemp, hemp processing, hemp grinding, homogenization, sample preparation, cannabinoid detection, THC, CBD
Background
Cannabis sativa, commonly known as “hemp,” has broad usages in industrial, medicinal, and agricultural applications across the world.[1] Currently, the applications of both hemp and marijuana are being explored in greater detail than previously seen by pharmaceutical and therapeutic applications.[2][3][4] The naturally occurring cannabinoid molecules found in hemp are the basis for its pharmacologic activity and viewed as a presumptive area of increasing importance in the field of pharmacology.[2][3] In addition to the medical application of cannabinoid-based compounds, the use of cannabinoid-infused products is on the rise for recreational usage.[5]
As states across the United States begin to legalize the sale and usage of marijuana and cannabinoid-derived products, there is an increased longing for products that will induce the reported psychoactive or therapeutic effects of these cannabinoids.[2][5][6] With demand for these products exponentially rising, the hemp industry has grown to a billion-dollar industry crossing recreational and medicinal markets for cannabinoids and cannabinoid-derived products.[5] However, as the industry has dramatically expanded in recent years, regulations and best-practice methods have not been keeping up.[6] Currently, there are a variety of unregulated techniques used in hemp processing for sample preparation and the assessment of products for cannabinoid concentrations.
Testing for the cannabinoids Δ9-tetrahydrocannabinolic acid (THCA) and cannabidiolic acid (CBDA), and their decarboxylated counterparts Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD), are critical in evaluating hemp-based products being brought to market.[7] In the U.S., hemp-based products must remain under certain concentrations of THCA and THC to remain labeled as hemp versus marijuana, a key determination in many areas given the legality of hemp versus marijuana in many municipalities.[8] Additionally, the CBDA and CBD concentrations within products have been related to their analgesic potency, thus accurate measurement and reporting of these values are vital to proper labeling and pricing of these products to protect both the supplier and consumer.[6][7][8]
Herein, we report a method for sample preparation of hemp involving bead-mill-based homogenization using a novel “active grinding media” that provides uniform processing of hemp samples with reproducible particle sizes and no alterations to the carboxylation states of THCA or CBDA. This method allows for accurate potency testing of hemp and hemp-based products through a homogenization that results in a completely processed, homogenous sample.
Methods
Sample preparation and homogenization
To measure cannabinoid potency, commercially available hemp was purchased. Hemp flower was placed in a 50 mL conical tube (Omni International, Cat. No. 19–6650), followed by the placement of the active grinding media (Omni International, Cat. No. 19-900 M) specifically for hemp. Tubes were loaded into the BR96 homogenizer (Omni International, Cat. No. 27–0001) in an even parallel series (2, 4, 6, or 8 tubes). Once loaded, the BR96 homogenizer was operated at 25 Hz for one cycle (15 seconds), two separate cycles (total of 30 seconds), four separate cycles (total of 60 seconds), and six separate cycles (total of 90 seconds).
Sieve analysis
A diamond tap sieves shaker (Esslinger.com, Cat. No. 51.0569) was used. Tap sieve shakers are specified in various standards for particle size analysis. The shaker was used in a horizontal and circular motion, and super-imposed by a vertical motion while administering a taping action. Tap sieve shakers was specified for specific particle size analysis (~ 1.10 mm to 4.50 mm). Starting weight before and after analysis was recorded, and distribution percentages were calculated.
Cannabinoid sample analysis for potency
Approximately 100 mg of homogenized flower matrix was isolated and 5 mL of methanol was added. Sample was sonicated, vortexed, and centrifugated. Supernatant was diluted with MeOH/H2O (80/20, v/v) + 0.1% formic acid as appropriate for injection. Sample was injected (5 μl) into the Agilent 1220 Infinity II LC. The analytical column used was the Infinity Poroshell 120 EC-C18, 3.0 mm × 50 mm, 2.7 μM at 50 °C. Gradient used was 0 to 1 minute, 60% B, 7–8.2 minutes, 60 to 77% B, and 8.2 to 10 minutes, 95% B.
Cannabinoid standards
Standards for THC, THCA, CBD, and CBDA (1 mg/ml) were obtained from Cerilliant (Round Rock, Texas). Standards were combined, and a stock solution with a concentration of 250 ppm for each cannabinoid was made and serially diluted in 100% methanol following the dilution series (Table 1). Fifteen μL of each dilution was diluted further with 35 μLs of 100% methanol and analyzed by reverse phase high-performance liquid chromatography (HPLC).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sample preparation
The six (1 mL samples of the 250-ppm cannabinoid) standards were added to reinforced tubes (2 mL) that were pre-filled with 2.8 mm ceramic beads (Omni International, Kennesaw, GA, Cat. No. 19–628). The tubes were processed on the Bead Ruptor Elite Bead Mill Homogenizer at 5 m/s for increasing durations of 10, 20, 30, 60, and 120 seconds. After each time point, 250 µL was removed to a new 1.5 mL microcentrifuge tube and placed on ice. As a positive control, 500 µL of the 250-ppm cannabinoid standard was placed in a 1.5 mL microcentrifuge tube and heated, at 90 °C, for 180 minutes. All samples were filtered through a 0.2 µm spin filter and 15 µL of the filtrate was combined with 35 µL of 100% HPLC grade methanol for reverse phase HPLC analysis.
Reverse phase HPLC for standards
Cannabinoid separation and quantification was performed on a Waters 1525 HPLC (Waters Corp, Milford, MA) equipped with a binary pump and 2996 photodiode array detector. Buffer A consisted of ddH2O and formic acid (0.2% v/v), and buffer B was Acetonitrile and formic acid (0.2% v/v). Fifteen μL of each sample was separated on a Raptor ARC-18 150 mm × 4.6 mm, 2.7 μm column over a 20-minute linear gradient from 60% B to 100% B at a flow rate of 1.5 mL/minute. Absorbance was measured at 280 nm.
Results
Through the homogenization methods described above, we have shown that our methodology results in homogenate particle sizes < 1.25 mm. Running one cycle of our optimized homogenization parameters yielded 11.2% of particles < 1.25 mm; we observed that with each additional cycle of 15 seconds of homogenization, we were able to increase the percentage of homogenate particles < 1.25 mm to as high as 60.2% of all particles within the sample after four cycles of 15 seconds (Table 1). This data suggests that intermittent homogenization produces a complete homogenate, with the average particle size decreasing after each cycle. As average particle size decreased with each cycle of homogenization, we also observed an increased extraction of CBD, CDBA, THC, and THCA, with a significant increase in the detection of CBD and CBDA (Table 2). These findings are depicted in the resulting average concentration of each cannabinoid and as the relative extraction efficiency, examining the difference in recovered cannabinoids after each homogenization parameter when compared to Set-1.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
In addition to providing particle sizes < 1.25 mm, the use of our active grinding media and cyclical homogenization approach did not cause decarboxylation changes to the THCA and CBDA populations seen in the hemp tested, as confirmed by reverse phase HPLC (Fig. 1, Supplementary File 1). Processing hemp samples under these parameters did not produce enough thermal energy to alter the carboxylation start of the major cannabinoids tested for. Maintaining temperatures during sample processing under 90 °C is the benchmark to ensure that cannabinoids being tested for downstream do not have their concentrations altered during processing via decarboxylation reactions (Fig. 2). Additional homogenization runs with extended run times, up to 180 seconds, were also run in an attempt to determine the processing time required to generate enough heat to induce the decarboxylation reactions that convert THCA to THC and CBDA to CBD (Fig. 1, Supplemental Fig. 1). These extended time homogenizations showed that our method maintains processing temperatures under 35 °C, when measuring the temperature both internal and external to tubes used for the sample processing throughout the run (Fig. 3).
|
|
|
Discussion
The method proposed in this manuscript for hemp sample preparation via homogenization with our novel active grinding media demonstrates a critical step forward in the commercial and medical hemp markets. This methodology brings forward a standardized protocol for hemp processing that allows for reproducible results, small particle sizes, and unaltered cannabinoid profiles, all of which are essential as the market expands and inevitably becomes more standardized and regulated. While it is no surprise that federal, state, and local regulations have not developed as rapidly as the market for hemp-based products has, we must be fully aware that these regulations are coming and work proactively to establish robust industry standard methods for processing and testing hemp-involved products.
The authors of this study firmly believe that this processing methodology is a strong first step towards establishing those industry standards for hemp processing and testing. Current guidelines surrounding hemp-derived products require that the measured THC levels remain below certain prescribed limits in order to remain labeled as hemp versus marijuana, a critical distinction in many markets across the United States where hemp is legal and marijuana is not.[6][8] While some argue that this is a game of legal semantics, this paradigm is critical for the producers of these products and compliance with these regulations is directly impacting their ability to remain in business. With such high stakes involved in accurate cannabinoid profiling, we were surprised to see so much variability in sample preparation techniques for these processes. The use of coffee grinders, blenders, and other various dissociation devices have two major problems seen when they are utilized for hemp sample preparation prior to cannabinoid profiling: 1. their dissociation processes are highly energetic, potentially heating the sample to a temperature to induce decarboxylation of THCA to THC, and 2. they are often very difficult to clean, allowing for cross contamination between samples.[9][10][11] Both of these are ameliorated with our proposed methodology, implying a superior process for sample preparation, robust enough to be implemented into a standardized regulatory workflow.
However, we do acknowledge that given the potential hundreds of coffee grinders and blenders in the current market which can be utilized for cannabis homogenization purposes, it would be impossible to compare each of them to our proposed methodology or even determine which is considered an “industry leader” brand in this market given the wide variability of techniques and lack of regulation. As such, we have elected to avoid direct comparisons with this proposed methodology. This purposeful decision of omission does not impact the utility of the proposed technology for hemp processing, but rather it further supports the need of a standardization of processing techniques to allow for future studies to benchmark against.
In addition to the critical need for a standardized process that will not impact the THC profile of a hemp sample, it also important that the CBD profile be measured accurately.[6][8] The CBD profile of a hemp-based product is often tied to the potency of the product, with higher concentrations of CBD fetching higher prices in the market. Ensuring that accurate measurement of these cannabinoids is done requires a process that does not induce excessive thermal energy to the sample and produces a reliably small particle size throughout the sample to ensure accurate measurement with HPLC.
As the hemp market continues to exponentially grow across the U.S., we feel that it is a pertinent time to adopt industry-standardized procedures. The significance of this work is to show that homogenization is a dependable and consistent method for measuring cannabinoid potency in hemp. With many states legally allowing hemp (Cannabis with less than 0.3% THC) usage but prohibiting the use of cannabis with high levels of THC, it is critical to develop processing technologies that will not alter these cannabinoid profiles. The conversion of THCA to active THC can redefine a hemp product, into a high THC cannabis product, rendering it illegal. Having an accurate measurement of all forms of cannabinoids is important to prevent this. We show that the smaller the particles of hemp the more accurate potency testing becomes. Furthermore, this method is repeatable, and no contamination from multiple samples occurs. Having accurate potency testing also helps growers market their premium product verses product of lesser grades of potency. The method described in this manuscript is a robust approach to hemp sample preparation which allows for accurate and reproducible downstream molecular analysis of each sample’s cannabinoid profile.
Conclusions
Herein, we are proposing a methodology for hemp sample preparation and homogenization that provides uniform homogenization of a sample, while maintaining the integrity and proportionality of the cannabinoids for downstream molecular analysis. Through use of the active grinding media and bead-mill homogenization, we prepared samples with a majority of the particles < 1.25 mm, resulting in increased cannabinoid recovery when compared to larger particle size homogenates, and no alterations in the carboxylation profiles of CBDA or THCA during processing. This method provides critical reproducibility to processing hemp, allowing the end user to maintain the highest of potency and purity standards when moving forward with their molecular analysis.
Supplementary information
- Supplementary File 1 (.docx)
Abbreviations
- THCA: Δ9-Tetrahydrocannabinolic acid
- THC: Tetrahydrocannabinol
- CBDA: Cannabidiolic acid
- CBD: Cannabidiol
- HPLC: High-performance liquid chromatography
Acknowledgements
The authors would like to acknowledge Karl Jahn, Pete Tortorelli, and Erik Masefield of Omni International, Inc. for their financial commitment and continued support of our research. Additionally, we would like to acknowledge Rachele Capobianco and Derick Gordon for their support in establishing the collaborations needed for this project. Finally, we would like to acknowledge Rachel True, Rachel Nash, and Leah Proctor for their sacrifices and unwavering support of our research. IANAFS.
Contributions
ZPM: formal analysis, writing—original draft, review and editing, visualization. GLR: investigation, formal analysis, writing—review and editing. CMP: methodology, investigation, writing—review and editing. AO: investigation. WT: methodology, investigation. DB: methodology, writing—review and editing, supervision, project administration. TW: investigation. SP: resources, supervision, funding acquisition. BM: investigation. VC investigation. BE: conceptualization, methodology, investigation, data curation, writing—reviewing and editing. JA: conceptualization, supervision, funding acquisition. RJN: conceptualization, investigation, resources, writing—original draft, review and editing, visualization, supervision, project administration, funding acquisition. YCP: conceptualization, investigation, resources, data curation, visualization, supervision, project administration.
Funding
This project was funded in its entirety by the private entities Omni International, Inc. and AMERICANNA Laboratories, LLC.
Availability of data and materials
All data used in the development of this manuscript is available through email requests to authors Z.P. Morehouse or R.J. Nash at zmorehouse at omni-inc dot com or rnash at omni-inc dot com respectively.
Conflict of interest
G.L. Ryan, C.M. Proctor, A. Okparanta, W. Todd, D. Bunting, T. White, B. Easparro, and J. Atwood are all employed by Omni International, Inc. but do not have any personal financial interests in the success of the company. Z.P. Morehouse has a consultancy contract with Omni International, Inc. but does not have any personal financial interests in the success of the company. R.J. Nash has a consultancy contract with Omni International, Inc. but does not have any personal financial interests in the success of the company, and is co-founder and owner of Jeevan Biosciences (Tucker, GA USA) to which he does have financial interests in its success. B. Miller, V. Colon, and Y.C. Park are all employed by AMERICANNA Laboratories, LLC and do not have any personal financial interests in its success. S. Perez is cofounder and co-owner of AMERICANNA Laboratories, LLC to which he does have personal financial interests in its success.
References
- ↑ Russo, Ethan B. (21 August 2007). "History of Cannabis and Its Preparations in Saga, Science, and Sobriquet" (in en). Chemistry & Biodiversity 4 (8): 1614–1648. doi:10.1002/cbdv.200790144. https://onlinelibrary.wiley.com/doi/10.1002/cbdv.200790144.
- ↑ 2.0 2.1 2.2 Devsi, Alykhan; Kiyota, Brett; Ouellette, Theophile; Hegle, Andrew P.; Rivera-Acevedo, Ricardo E.; Wong, Jasper; Dong, Ying; Pugsley, Michael K. et al. (1 December 2020). "A pharmacological characterization of Cannabis sativa chemovar extracts" (in en). Journal of Cannabis Research 2 (1): 17. doi:10.1186/s42238-020-00026-0. ISSN 2522-5782. PMC PMC7819338. PMID 33526117. https://jcannabisresearch.biomedcentral.com/articles/10.1186/s42238-020-00026-0.
- ↑ 3.0 3.1 Amin, Md Ruhul; Ali, Declan W. (2019), Bukiya, Anna N., ed., "Pharmacology of Medical Cannabis" (in en), Recent Advances in Cannabinoid Physiology and Pathology (Cham: Springer International Publishing) 1162: 151–165, doi:10.1007/978-3-030-21737-2_8, ISBN 978-3-030-21736-5, http://link.springer.com/10.1007/978-3-030-21737-2_8
- ↑ Ebbert, Jon O.; Scharf, Eugene L.; Hurt, Ryan T. (1 December 2018). "Medical Cannabis" (in en). Mayo Clinic Proceedings 93 (12): 1842–1847. doi:10.1016/j.mayocp.2018.09.005. https://linkinghub.elsevier.com/retrieve/pii/S0025619618307092.
- ↑ 5.0 5.1 5.2 Kumar, Navin; Puljević, Cheneal; Ferris, Jason; Winstock, Adam; Barratt, Monica J. (1 December 2019). "Cannabis use patterns at the dawn of US cannabis reform" (in en). Journal of Cannabis Research 1 (1): 5. doi:10.1186/s42238-019-0003-z. ISSN 2522-5782. PMC PMC7815050. PMID 33526080. https://jcannabisresearch.biomedcentral.com/articles/10.1186/s42238-019-0003-z.
- ↑ 6.0 6.1 6.2 6.3 6.4 Abuhasira, Ran; Shbiro, Liat; Landschaft, Yuval (1 March 2018). "Medical use of cannabis and cannabinoids containing products – Regulations in Europe and North America" (in en). European Journal of Internal Medicine 49: 2–6. doi:10.1016/j.ejim.2018.01.001. https://linkinghub.elsevier.com/retrieve/pii/S0953620518300013.
- ↑ 7.0 7.1 Coogan, Thomas A. (1 December 2019). "Analysis of the cannabinoid content of strains available in the New Jersey Medicinal Marijuana Program" (in en). Journal of Cannabis Research 1 (1): 11. doi:10.1186/s42238-019-0011-z. ISSN 2522-5782. PMC PMC7819311. PMID 33526081. https://jcannabisresearch.biomedcentral.com/articles/10.1186/s42238-019-0011-z.
- ↑ 8.0 8.1 8.2 8.3 Mead, Alice (1 May 2017). "The legal status of cannabis (marijuana) and cannabidiol (CBD) under U.S. law" (in en). Epilepsy & Behavior 70: 288–291. doi:10.1016/j.yebeh.2016.11.021. https://linkinghub.elsevier.com/retrieve/pii/S1525505016305856.
- ↑ Atkins, Patricia L (1 March 2019). "Sample Processing and Preparation Considerations for Solid Cannabis Products" (in en). Journal of AOAC INTERNATIONAL 102 (2): 427–433. doi:10.5740/jaoacint.18-0203. ISSN 1060-3271. https://academic.oup.com/jaoac/article/102/2/427/5658162.
- ↑ Pacifici, Roberta; Marchei, Emilia; Salvatore, Francesco; Guandalini, Luca; Busardò, Francesco Paolo; Pichini, Simona (28 January 2017). "Evaluation of cannabinoids concentration and stability in standardized preparations of cannabis tea and cannabis oil by ultra-high performance liquid chromatography tandem mass spectrometry". Clinical Chemistry and Laboratory Medicine (CCLM) 55 (10): 1555–1563. doi:10.1515/cclm-2016-1060. ISSN 1437-4331. https://www.degruyter.com/document/doi/10.1515/cclm-2016-1060/html.
- ↑ Ibrahim, Elsayed; Wang, Mei; Radwan, Mohamed; Wanas, Amira; Majumdar, Chandrani; Avula, Baharthi; Wang, Yan-Hong; Khan, Ikhlas et al. (1 March 2019). "Analysis of Terpenes in Cannabis sativa L. Using GC/MS: Method Development, Validation, and Application" (in en). Planta Medica 85 (05): 431–438. doi:10.1055/a-0828-8387. ISSN 0032-0943. http://www.thieme-connect.de/DOI/DOI?10.1055/a-0828-8387.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added. Nothing else was changed in accordance with the NoDerivatives portion of the license.