Difference between revisions of "Journal:Development of an electronic information system for the management of laboratory data of tuberculosis and atypical mycobacteria at the Pasteur Institute in Côte d’Ivoire"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Finished adding rest of content.) |
||
| Line 19: | Line 19: | ||
|download = [http://jhmi.sums.ac.ir/index.php/JHMI/article/download/513/160 http://jhmi.sums.ac.ir/index.php/JHMI/article/download/513/160] (PDF) | |download = [http://jhmi.sums.ac.ir/index.php/JHMI/article/download/513/160 http://jhmi.sums.ac.ir/index.php/JHMI/article/download/513/160] (PDF) | ||
}} | }} | ||
==Abstract== | ==Abstract== | ||
'''Introduction''': Tuberculosis remains a public health problem despite all the efforts made to eradicate it. To strengthen the surveillance system for this condition, it is necessary to have a good data management system. Indeed, the use of electronic information systems in [[Information management|data management]] can improve the quality of data. The objective of this project was to set up a laboratory-specific electronic information system for mycobacteria and atypical tuberculosis. | '''Introduction''': Tuberculosis remains a public health problem despite all the efforts made to eradicate it. To strengthen the surveillance system for this condition, it is necessary to have a good data management system. Indeed, the use of electronic information systems in [[Information management|data management]] can improve the quality of data. The objective of this project was to set up a laboratory-specific electronic information system for mycobacteria and atypical tuberculosis. | ||
| Line 51: | Line 47: | ||
===Architecture and system features=== | ===Architecture and system features=== | ||
We opted for a web-based application used in the company’s intranet. It should be noted that the project was entirely based on Java technology. The design of the application was based on the Apache Struts 2.0 framework<ref name="ApacheStruts">{{cite web |url=https://struts.apache.org/ |title=Apache Struts |publisher=Apache Software Foundation |accessdate=04 October 2017}}</ref>, which defined a controller-view-template three-layer design. The database connection layer uses Hibernate<ref name="Hibernate">{{cite web |url=http:// | We opted for a web-based application used in the company’s intranet. It should be noted that the project was entirely based on Java technology. The design of the application was based on the Apache Struts 2.0 framework<ref name="ApacheStruts">{{cite web |url=https://struts.apache.org/ |title=Apache Struts |publisher=Apache Software Foundation |accessdate=04 October 2017}}</ref>, which defined a controller-view-template three-layer design. The database connection layer uses Hibernate<ref name="Hibernate">{{cite web |url=http://hibernate.org/ |title=Hibernate |publisher=Red Hat, Inc |date=04 May 2018}}</ref>, which is an object-relational mapping (ORM) open-source solution that facilitates the development of the persistence layer; this database connection layer is also based on Java Database Connectivity (JDBC) for some modules of the application. The production of PDF reports was possible by the use of the JasperReports Library.<ref name="JRLib">{{cite web |url=https://community.jaspersoft.com/project/jasperreports-library |title=JasperReports Library |publisher=TIBCO Software, Inc |accessdate=16 January 2018}}</ref> The reports in the Excel format required the libraries of the Apache POI project.<ref name="ApachePOI">{{cite web |title=Apache POI |publisher=Apache Software Foundation |accessdate=10 May 2018}}</ref> The Java Enterprise Edition of Glassfish 4.1<ref name="GlassFish">{{cite web |url=https://javaee.github.io/glassfish |title=GlassFish: The Open Source Java EE Reference Implementation |author=Oracle Corporation |work=Github |accessdate=14 January 2019}}</ref> deployment server in its free version was used to host the application, and the [[MySQL]] 5.1 database management system (DBMS) was used to manage the database. The MySQL DBMS and Glassfish were installed on an HP ProLiant server running on a UNIX CentOS 6.7 OS. All the client machines were PC-type computers. All web browsers could be used as web clients for the application, though with a preference for Google Chrome; browsers such as Internet Explorer, Safari, and Firefox do not fully support the “date” and “time” input types.<ref name="PatreonDateAndTime">{{cite web |url=https://caniuse.com/#feat=input-datetime |title=Date and time input types |work=Can I use |publisher=Patreon |accessdate=14 January 2019}}</ref> The data stored in the database benefited from automatic backups to an external hard drive, and replication of this external hard drive was done at the end of the week. The source code of the application as well as the different steps of the implementation was documented to allow future maintenance. The user interface was modeled to be fairly similar to the original paper form. System access management was also taken into account in the implementation. The system modules are accessible according to the rights assigned by the administrator. | ||
hibernate.org/ |title=Hibernate |publisher=Red Hat, Inc |date=04 May 2018}}</ref>, which is an object-relational mapping (ORM) open-source solution that facilitates the development of the persistence layer; this database connection layer is also based on Java Database Connectivity (JDBC) for some modules of the application. The production of PDF reports was possible by the use of the JasperReports Library.<ref name="JRLib">{{cite web |url=https://community.jaspersoft.com/project/jasperreports-library |title=JasperReports Library |publisher=TIBCO Software, Inc |accessdate=16 January 2018}}</ref> The reports in the Excel format required the libraries of the Apache POI project.<ref name="ApachePOI">{{cite web |title=Apache POI |publisher=Apache Software Foundation |accessdate=10 May 2018}}</ref> The Java Enterprise Edition of Glassfish 4.1<ref name="GlassFish">{{cite web |url=https://javaee.github.io/glassfish |title=GlassFish: The Open Source Java EE Reference Implementation |author=Oracle Corporation |work=Github |accessdate=14 January 2019}}</ref> deployment server in its free version was used to host the application, and the [[MySQL]] 5.1 database management system (DBMS) was used to manage the database. The MySQL DBMS and Glassfish were installed on an HP ProLiant server running on a UNIX CentOS 6.7 OS. All the client machines were PC-type computers. All web browsers could be used as web clients for the application, though with a preference for Google Chrome; browsers such as Internet Explorer, Safari, and Firefox do not fully support the “date” and “time” input types.<ref name="PatreonDateAndTime">{{cite web |url=https://caniuse.com/#feat=input-datetime |title=Date and time input types |work=Can I use |publisher=Patreon |accessdate=14 January 2019}}</ref> The data stored in the database benefited from automatic backups to an external hard drive, and replication of this external hard drive was done at the end of the week. The source code of the application as well as the different steps of the implementation was documented to allow future maintenance. The user interface was modeled to be fairly similar to the original paper form. System access management was also taken into account in the implementation. The system modules are accessible according to the rights assigned by the administrator. | |||
===On-site system installation and user training=== | ===On-site system installation and user training=== | ||
| Line 141: | Line 136: | ||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Strongly Agree | ! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Strongly Agree | ||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Agree | ! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Agree | ||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Neither Agree | ! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Neither Agree<br />Nor Disagree | ||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Disagree | ! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Disagree | ||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Strongly Disagree | ! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Strongly Disagree | ||
| Line 183: | Line 178: | ||
|} | |} | ||
==Discussion== | |||
We have developed an electronic laboratory information system that takes the specifics of a microbiology laboratory focused on mycobacteria into account. This LIS, based on the existing paper collection form, has been implemented to manage the data resulting from the activities of this laboratory, except for the management of laboratory inputs. | |||
The workflow begins with the sample arrival in the laboratory for examination of the bulletins coupled to billing. The invoicing forms attest to the agreement of the office of entries to the processing of the application by the laboratory. From the information on the request for examination, a search is made on the last name and name of the patient in the LIS at the search field of the page “Create new patient record.“ If there is ambiguity for a name—e.g, say that we find several identical names and surnames—other parameters such as date of birth, date of registration, telephone number, or residential city can be used to discriminate the patients and find the one that corresponds to the patient on the examination form. In case there is no match, a new folder is created, and the identification number used for this electronic file is generated manually from the laboratory register. This step of the process is often a bottleneck because people can have the same names and first names; the search can then become long. A unique identification number generated from the laboratory’s office of entries would solve this problem, but it requires re-engineering of the procedures of the office of entries. | |||
After sample processing, the corresponding patient record previously created is searched from the sample number or name and then opened. The visit form is created and all the results obtained are recorded. Thus, for the same patient, the LIS makes it possible to follow the evolution of its biological parameters from its visit forms. The LIS has saved time in the process of generating results before the patients results were enrolled manually on a pre-print form made in Microsoft Word. This process required at least five minutes per result; the generation time was less than a minute with the LIS.<ref name="El-KarehImpact12" /> | |||
To this day, all the results of LMTA come from the LIS; this has the spirit of improving the completeness of the data recorded. Pre-recorded drop-down lists, formatted fields such as the date field, and controls set up on free fields such as sample number fields reduce the recording of erroneous data and consequently improve their quality, but there are errors inherent in the user that the system cannot control such as the registration error on a patient’s name or age. | |||
Dashboards play an important role in monitoring laboratory activities and checking the evolution of the population health status. They also help monitor the laboratory’s performance indicator and the drug resistance shown by the MDR-TB and XDR-TB. | |||
Overall, the LIS was very well accepted, and all the staff of the laboratory found it comfortable since it is commonly used in the laboratory’s data management activities. The figures of the survey carried out is in the same line with those of the study carried out in South Africa which showed the good perception of computer systems in the management of health structures.<ref name="ClineInform13">{{cite journal |title=Information technology systems in public sector health facilities in developing countries: the case of South Africa |journal=BMC Medical Informatics and Decision Making |author=Cline, G.B.; Luiz, J.M. |volume=13 |pages=13 |year=2013 |doi=10.1186/1472-6947-13-13 |pmid=23347433 |pmc=PMC3570341}}</ref> A lab technician disagreed with the statement that the system saves time in performing data management tasks. After investigation, some bugs were found during the generation of the smear distribution table depending on the result, type of the sample, origin of the biological product, and the profile of the sample, thus motivating this response. This particular malfunction has been corrected. Unlike the study conducted by Barbara Castelnuovo ''et al.''<ref name="CastelnuovoImp12" /> who showed unwilling users adopted the electronic information systems, our study demonstrated the total approval of the laboratory staff as to upgrading to the LIS, as shown by the results of the LIS evaluation by users. | |||
==Conclusions== | |||
The introduction of the information system for the microbiology laboratory has necessitated the understanding and modeling of various processes used from the arrival of the examination requests to the printing of the results. The system, currently in use, has been readily adopted by the laboratory staff, who see a tool facilitating their work for laboratory technicians. LIS reduces the workload by automating a generation of the results and reports. For the biologists in charge of the laboratory, the LIS—through the dashboards—allows them to have in real time the indicators on the follow-up of samples, the activity carried out in the laboratory, and the state of resistance to antituberculosis treatments. The LIS has a positive impact on the laboratory activities, but obtaining a perfect tool is only done in a cycle of sustained improvement. | |||
==Additional material== | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="100%" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" colspan="6"|'''Appendix A''' Questionnaire Instrument (Likert Scale Structured Questions)<br />Please answer the following questions by indicating which answer most accurately represents the extent to which you agree or disagree with the statement on the left. There can only be one answer per statement. | |||
|- | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"| | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Strongly Agree | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Agree | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Neither Agree<br />Nor Disagree | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Disagree | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Strongly Disagree | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1. I am completely comfortable with LIS | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2. LIS saves time in completing tasks | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|3. The LIS allows access to information quickly and securely | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|4. LIS improves the overall quality of work | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|5. I plan to move to the exclusive use of the LIS for laboratory data management | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
|- | |||
|} | |||
|} | |||
==Acknowledgements== | |||
The authors would like to thank all the members of the Epidemiology and Laboratory Unit for Tuberculous and Atypical Mycobacteria. Special thanks to Anatole Mian for the English translation. | |||
===Funding=== | |||
This work did not receive any internal funding for its design. This is a project of the Epidemiology Unit to improve the data management of the National Reference Center for Tuberculosis in Côte d’Ivoire. No external funding was also received for this project. | |||
===Contributions of the authors=== | |||
CJK made the modeling and design, coding the application and writing the manuscript, TA made the conception and modeling of the Fact Sheet, MKD reading and validation of the manuscript, NR made the conception and validation of graphical interfaces and dashboard, SMK conception and modelisation epidemiological record reading and validation of the manuscript. All authors read and approved the final manuscript. | |||
===Conflict of interest=== | |||
None declared. | |||
==References== | ==References== | ||
| Line 188: | Line 262: | ||
==Notes== | ==Notes== | ||
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. We also added PMCID and DOI when they were missing from the original reference. Under "Architecture and system features," the original article had a poorly punctuated and confusing sentence about browser support for date and time input types; it has been updated to be more correct, with a citation added. The URL to GlassFish was updated to show the 4.x versions. | This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. We also added PMCID and DOI when they were missing from the original reference. Under "Architecture and system features," the original article had a poorly punctuated and confusing sentence about browser support for date and time input types; it has been updated to be more correct, with a citation added. The URL to GlassFish was updated to show the 4.x versions. The original had reference 13 in the References section, but it was never referenced in-line; it was omitted in this version. | ||
<!--Place all category tags here--> | <!--Place all category tags here--> | ||
Latest revision as of 23:31, 14 January 2019
| Full article title | Development of an electronic information system for the management of laboratory data of tuberculosis and atypical mycobacteria at the Pasteur Institute in Côte d’Ivoire |
|---|---|
| Journal | Journal of Health Management and Informatics |
| Author(s) | Koné, Constant J.; Touré, Assata; N’Dri, Mathias K.; Nguessan, Raymond; Soumahoro, Man-Koumba |
| Author affiliation(s) | Pasteur Institute of Côte d’Ivoire |
| Primary contact | Email: koneconstant at pasteur dot ci |
| Year published | 2019 |
| Volume and issue | 6(1) |
| Page(s) | 1–6 |
| DOI | None |
| ISSN | 2423-5857 |
| Distribution license | Creative Commons Attribution 3.0 Unported |
| Website | http://jhmi.sums.ac.ir/index.php/JHMI/article/view/513/160 |
| Download | http://jhmi.sums.ac.ir/index.php/JHMI/article/download/513/160 (PDF) |
Abstract
Introduction: Tuberculosis remains a public health problem despite all the efforts made to eradicate it. To strengthen the surveillance system for this condition, it is necessary to have a good data management system. Indeed, the use of electronic information systems in data management can improve the quality of data. The objective of this project was to set up a laboratory-specific electronic information system for mycobacteria and atypical tuberculosis.
Methods: The design of this laboratory information system required a general understanding of the workflow and the implementation processes in order to generate a realistic model. For the implementation of the system, Java technology was used to develop a web application compatible with the intranet of the company. The impact and the acceptability of the use of the system on the running of the laboratory were evaluated using the Likert scale.
Results: The system in place has been in operation for about 12 months, in conjunction with the paper registers. Since then, 4811 requests for examinations concerning 6083 samples have been registered. The results of analysis of 3892 patients were printed from the laboratory information system. In order to produce tuberculosis drug resistance reports and laboratory performance reports, dashboards have been developed.
Conclusion: The system has been adopted by the staff because of the time and efficiency gained in managing laboratory data. However, obtaining an optimized tool will only be done in a cycle of sustained improvement.
Keywords: clinical laboratory information systems, public health, tuberculosis
Introduction
Tuberculosis remains a public health problem despite all efforts to eradicate it. According to the World Health Organization (WHO), the number of tuberculosis cases in the world was estimated 9.6 million in 2014.[1] To strengthen the surveillance system for this disease, we need to have a good data management system.
The use of electronic information systems can improve the quality of clinical data[2] and thus the management of patients. Studies have shown that the implementation of computer tools is a performance factor for laboratory activities.[3] The results of laboratory analyses provide important data for clinical decision-making and treatment. Effective management of this data is essential for strengthening the health care system as a whole.
Laboratory information systems have been developed and used for about half a century.[3][4] Like all laboratory information systems, the microbiology laboratory information system must be secure, user-friendly, and able to interact with other information systems. However, there are several unique features of microbiology that are not used in other clinical laboratories, i.e., tracking multiple drifts, laboratory electronic notes, reporting results, and taking the preliminary and final results into account.[3]
The strategic plan of development of e-health has been defined and is being implemented.[5] All health facilities have to implement an electronic information system for better management of data generated by their activities.
The purpose of this project was to set up a laboratory information system (LIS) at Institute Pasteur of Côte d’Ivoire to collect specific data for Mycobacterium tuberculosis. This LIS will allow the recording of the data resulting from the activities of the laboratory. It should also allow for the generation of results that are ultimately given to the patient as well as the generation of reports on the activities of the laboratory. Finally, an analysis of the impact of this new system in the laboratory routine was carried out.
Methods
Design
Designing a laboratory information system requires an understanding of the workflow and the processes implemented. The data collection and recording sheet was the starting point for this work as the first objective, which was to build a system capable of recording the data and results from the laboratory’s activities. This data sheet is a key element in the activities of the laboratory since all patient information such as socio-demographic data, information on the samples, the preliminary and final results, and the notes of the various examinations are notified. This sheet was previously reorganized by the epidemiology unit. Then, from this new form and a series of interviews with biologists, some improvements were made. It was noted that in the paper recording system used before, the biological monitoring of a patient throughout his/her treatment was not possible. This feature was thus integrated into the new system. After analysis of all data, an application with conceptual, logic, and physical characteristics was designed using the Merise method in the open source tool MySQL Workbench.[6][7]
Architecture and system features
We opted for a web-based application used in the company’s intranet. It should be noted that the project was entirely based on Java technology. The design of the application was based on the Apache Struts 2.0 framework[8], which defined a controller-view-template three-layer design. The database connection layer uses Hibernate[9], which is an object-relational mapping (ORM) open-source solution that facilitates the development of the persistence layer; this database connection layer is also based on Java Database Connectivity (JDBC) for some modules of the application. The production of PDF reports was possible by the use of the JasperReports Library.[10] The reports in the Excel format required the libraries of the Apache POI project.[11] The Java Enterprise Edition of Glassfish 4.1[12] deployment server in its free version was used to host the application, and the MySQL 5.1 database management system (DBMS) was used to manage the database. The MySQL DBMS and Glassfish were installed on an HP ProLiant server running on a UNIX CentOS 6.7 OS. All the client machines were PC-type computers. All web browsers could be used as web clients for the application, though with a preference for Google Chrome; browsers such as Internet Explorer, Safari, and Firefox do not fully support the “date” and “time” input types.[13] The data stored in the database benefited from automatic backups to an external hard drive, and replication of this external hard drive was done at the end of the week. The source code of the application as well as the different steps of the implementation was documented to allow future maintenance. The user interface was modeled to be fairly similar to the original paper form. System access management was also taken into account in the implementation. The system modules are accessible according to the rights assigned by the administrator.
On-site system installation and user training
The server housing the executable files of the information system and the database was installed in a room which was secure and protected from the weather. A high capacity UPS was connected to this server to prevent the system from shutting down in the event of a power outage. The client computers comprised the ordinary service computers. The application was available on the corporate intranet from a web browser.
Training of six members of the laboratory staff was conducted in half a day. The assimilation took place quite quickly because the members of the laboratory all had good notions of the computer tool. Consultations were often sought when certain aspects had been forgotten when using the system. After each update, a short information session was organized to share the new features, after which use of the software continued without interruption. Biologists' use of the LIS often resulted in the discovery of bugs, which were later corrected; suggestions and comments were often made by users to improve this information system.
Qualitative assessment
The management of patients with tuberculosis involves regular biological tests. Monitoring of the analyses over time for a patient becomes problematic, which is also the case with the search for information on a sample, as well as the completion of activity reports and the delivery of results. The information system put in place should be able to improve the realization of these tasks on a daily basis.
After 12 months of usage, we evaluated the acceptability of this information system and the impact of implementation on the functioning of the laboratory by different users of the system, namely a data manager, two technicians, two engineers and a biologist who had to use the software continuously. This evaluation was based on the Likert scale.[14] To do this, we administered a questionnaire to selected the staff members. The interviewees were to choose one response from the five possible responses given for each of the questions which included: Strongly Agree (5), Agree (4), Neither Agree or Disagree (3), Disagree (2), and Strongly Disagree (1). Summary statistics were generated from the responses obtained.
Results
Results of the system launch
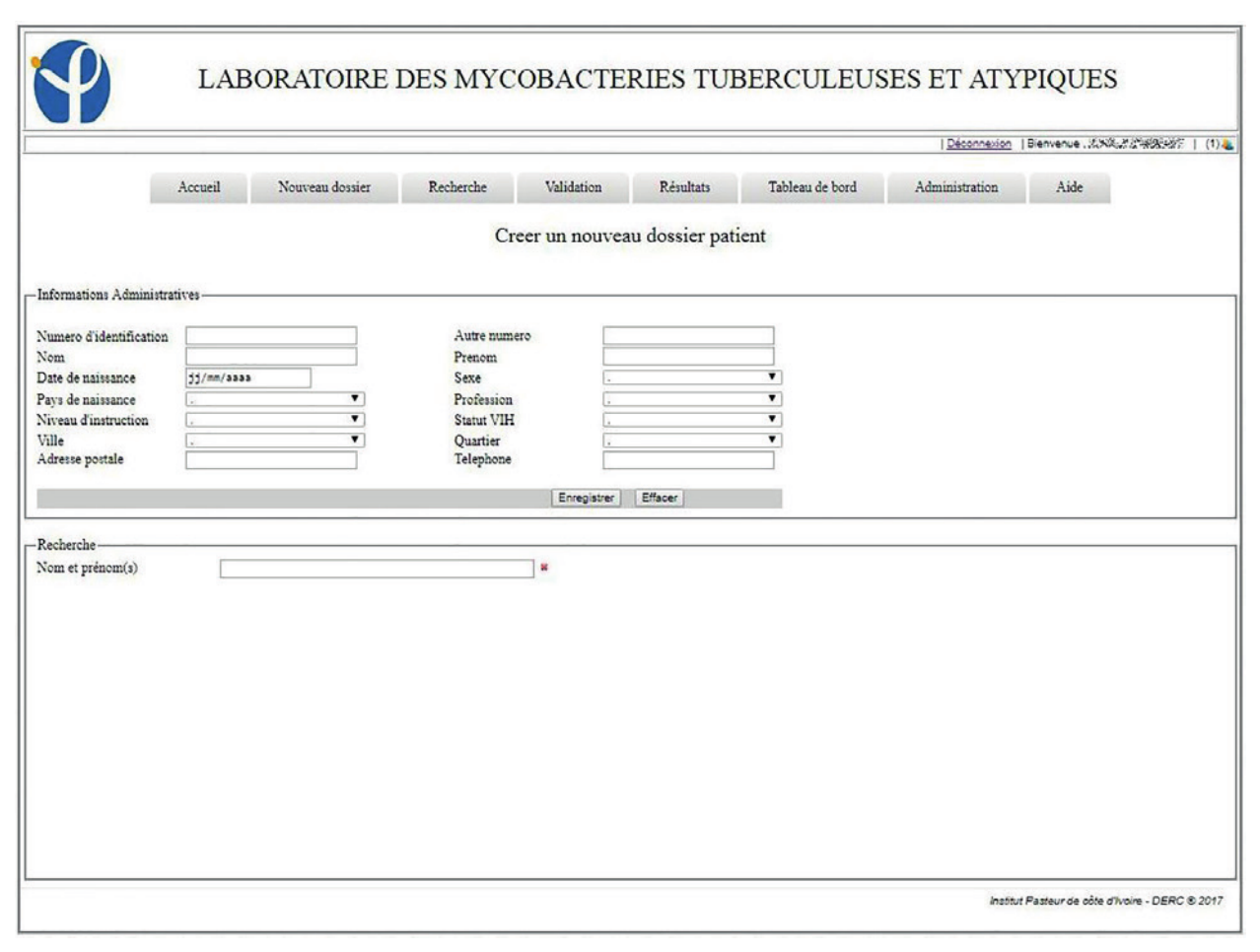
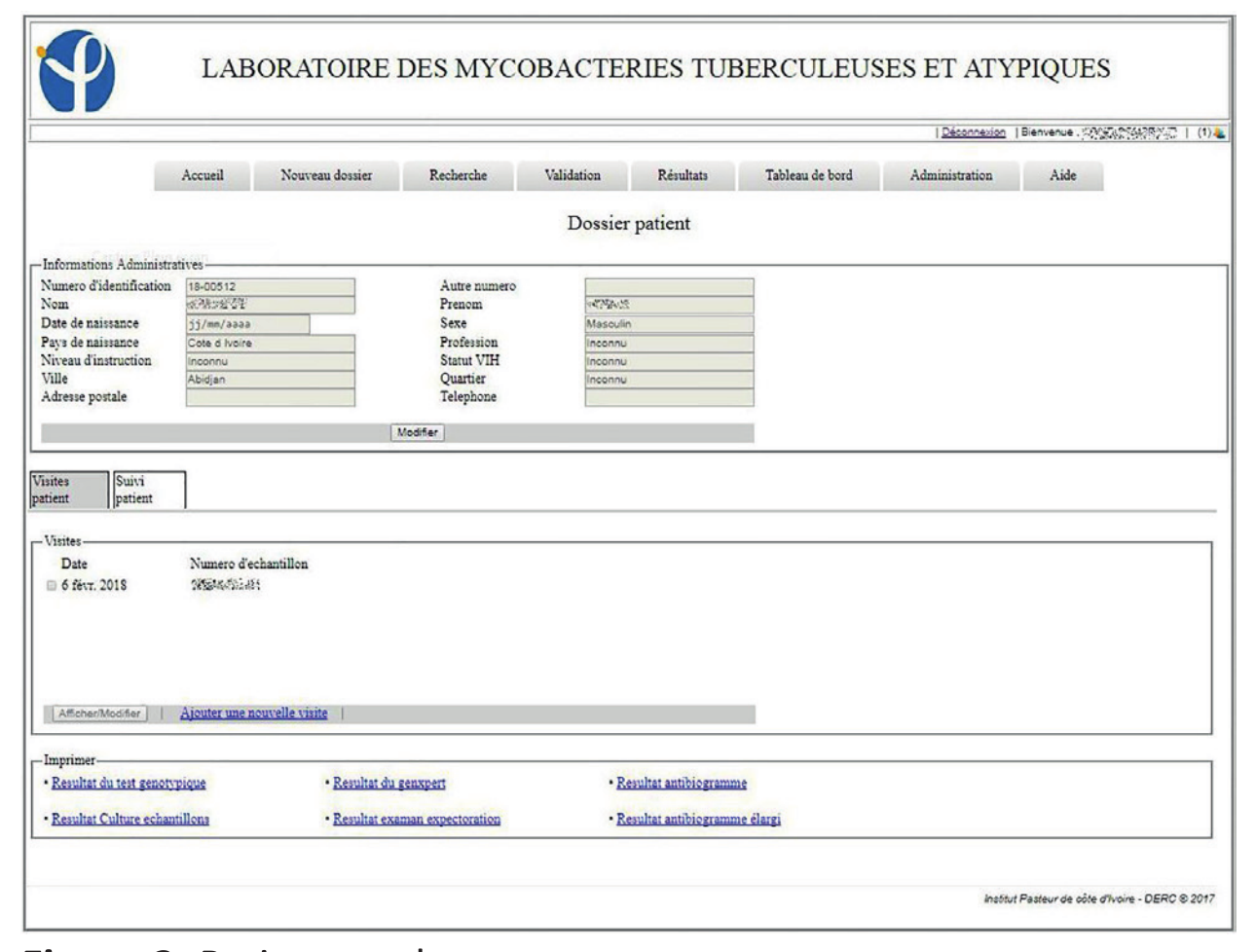
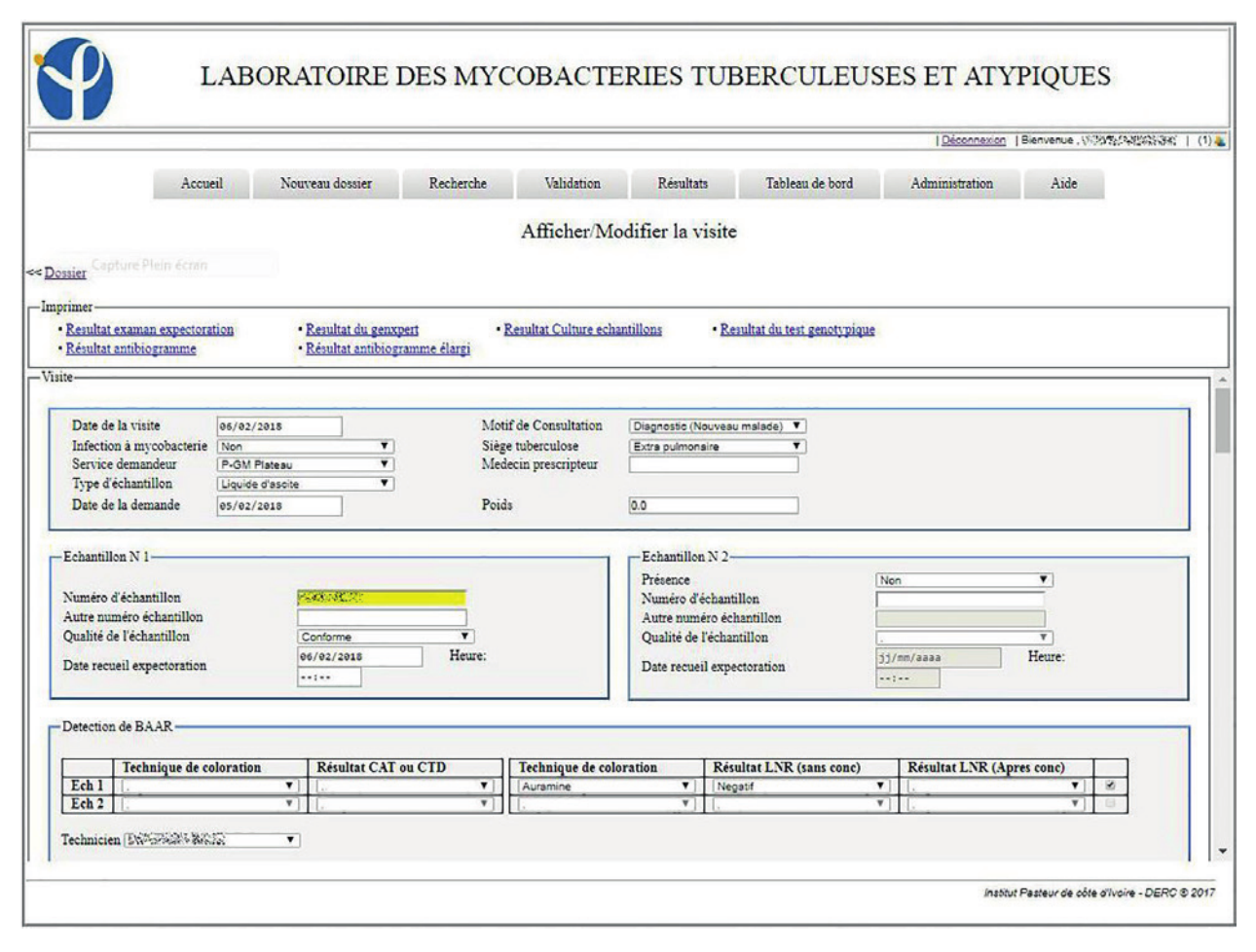
The system in place has been in use since the January 1, 2017. By the end of the year, 3,892 patients were registered in the LIS, with 4,811 visits and 6,083 collected samples recorded. Of the registered patients, 805 made at least two visits to the laboratory. In this information system, 93.9% (185/197) of the entry fields from patient and visit cards are fields with drop-down lists of those pre-recorded at the level of the administration module. These mechanisms were put in place to avoid the errors inherent in free text field filling. LIS allowed the patient folder creation as well as the creation of visit cards related to requests for examinations. These visit sheets were linked to the same record for a patient, which helps to monitor his/her condition (Figures 1, 2, 3).
|
|
|
The printing of the results intended for the patients requires preliminary validation by rhe biologist responsible for test results; otherwise, the request for printing is not executed. Six laboratory-delivered results could be printed, including microscopy, Genexpert, solid and/or fluid culture, genotypic test, standard antibiogram, and extended antibiogram. In the interests of quality assurance of the laboratory, the second impression for the same result carries the duplicate mark, so during this period, 6,075 printed results were obtained, which included 3,415 microscopy examinations, 1,591 Genexpert and LPA tests, 35 culture samples, 90 classical antibiogram, and five expanded.
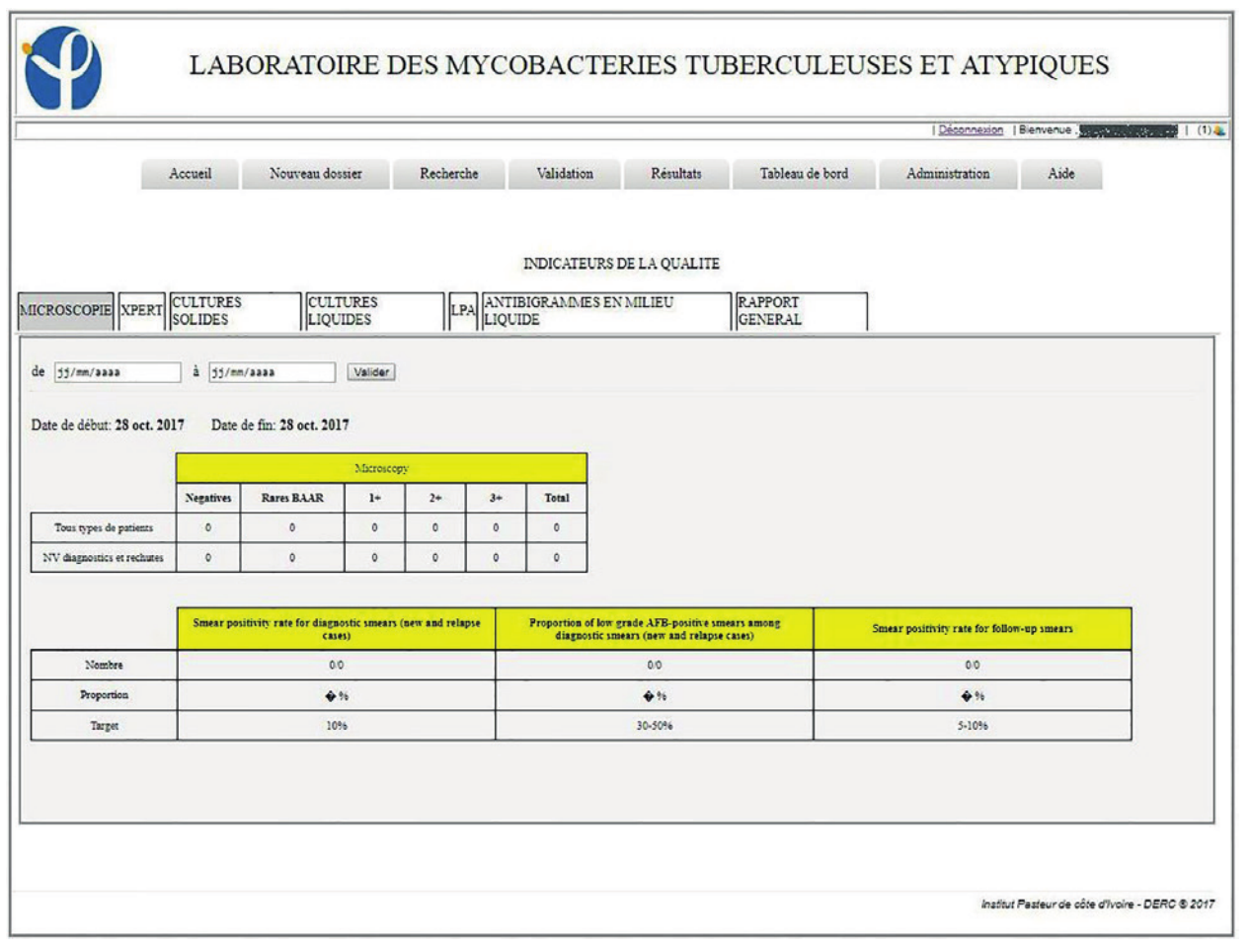
A dashboard system allows the calculation of indicators for the monitoring of laboratory activities and also provides indicators on the state of the health of the patients. These dashboards generate indicators for activities, including microscopy, Genexpert, culture in liquids and solids media, LPA, and antibiotic sensitivity (antibiogram) for a period defined by a time interval to complete. It also allows you to generate an Excel spreadsheet, the list of samples with associated information such as sample number, methods, and test results (Figures 4, 5).
|
|
Perception and usability of the system
The questionnaire was administered to the laboratory staff. Table 1 details the results obtained for these five questions. Regarding the assertions, they’re saying “the user feels comfortable with the LIS,” “LIS allows security and quick access to information,” “the LIS improves the quality of the work in the laboratory,” and “the user considers to switch to exclusive use of the electronics for managing the laboratory data”; 100% (6/6) responded favorably. One in six found that LIS did not save time in performing data management tasks.
| ||||||||||||||||||||||||||||||||||||||||||
Discussion
We have developed an electronic laboratory information system that takes the specifics of a microbiology laboratory focused on mycobacteria into account. This LIS, based on the existing paper collection form, has been implemented to manage the data resulting from the activities of this laboratory, except for the management of laboratory inputs.
The workflow begins with the sample arrival in the laboratory for examination of the bulletins coupled to billing. The invoicing forms attest to the agreement of the office of entries to the processing of the application by the laboratory. From the information on the request for examination, a search is made on the last name and name of the patient in the LIS at the search field of the page “Create new patient record.“ If there is ambiguity for a name—e.g, say that we find several identical names and surnames—other parameters such as date of birth, date of registration, telephone number, or residential city can be used to discriminate the patients and find the one that corresponds to the patient on the examination form. In case there is no match, a new folder is created, and the identification number used for this electronic file is generated manually from the laboratory register. This step of the process is often a bottleneck because people can have the same names and first names; the search can then become long. A unique identification number generated from the laboratory’s office of entries would solve this problem, but it requires re-engineering of the procedures of the office of entries.
After sample processing, the corresponding patient record previously created is searched from the sample number or name and then opened. The visit form is created and all the results obtained are recorded. Thus, for the same patient, the LIS makes it possible to follow the evolution of its biological parameters from its visit forms. The LIS has saved time in the process of generating results before the patients results were enrolled manually on a pre-print form made in Microsoft Word. This process required at least five minutes per result; the generation time was less than a minute with the LIS.[4]
To this day, all the results of LMTA come from the LIS; this has the spirit of improving the completeness of the data recorded. Pre-recorded drop-down lists, formatted fields such as the date field, and controls set up on free fields such as sample number fields reduce the recording of erroneous data and consequently improve their quality, but there are errors inherent in the user that the system cannot control such as the registration error on a patient’s name or age.
Dashboards play an important role in monitoring laboratory activities and checking the evolution of the population health status. They also help monitor the laboratory’s performance indicator and the drug resistance shown by the MDR-TB and XDR-TB.
Overall, the LIS was very well accepted, and all the staff of the laboratory found it comfortable since it is commonly used in the laboratory’s data management activities. The figures of the survey carried out is in the same line with those of the study carried out in South Africa which showed the good perception of computer systems in the management of health structures.[15] A lab technician disagreed with the statement that the system saves time in performing data management tasks. After investigation, some bugs were found during the generation of the smear distribution table depending on the result, type of the sample, origin of the biological product, and the profile of the sample, thus motivating this response. This particular malfunction has been corrected. Unlike the study conducted by Barbara Castelnuovo et al.[2] who showed unwilling users adopted the electronic information systems, our study demonstrated the total approval of the laboratory staff as to upgrading to the LIS, as shown by the results of the LIS evaluation by users.
Conclusions
The introduction of the information system for the microbiology laboratory has necessitated the understanding and modeling of various processes used from the arrival of the examination requests to the printing of the results. The system, currently in use, has been readily adopted by the laboratory staff, who see a tool facilitating their work for laboratory technicians. LIS reduces the workload by automating a generation of the results and reports. For the biologists in charge of the laboratory, the LIS—through the dashboards—allows them to have in real time the indicators on the follow-up of samples, the activity carried out in the laboratory, and the state of resistance to antituberculosis treatments. The LIS has a positive impact on the laboratory activities, but obtaining a perfect tool is only done in a cycle of sustained improvement.
Additional material
| ||||||||||||||||||||||||||||||||||||||||||
Acknowledgements
The authors would like to thank all the members of the Epidemiology and Laboratory Unit for Tuberculous and Atypical Mycobacteria. Special thanks to Anatole Mian for the English translation.
Funding
This work did not receive any internal funding for its design. This is a project of the Epidemiology Unit to improve the data management of the National Reference Center for Tuberculosis in Côte d’Ivoire. No external funding was also received for this project.
Contributions of the authors
CJK made the modeling and design, coding the application and writing the manuscript, TA made the conception and modeling of the Fact Sheet, MKD reading and validation of the manuscript, NR made the conception and validation of graphical interfaces and dashboard, SMK conception and modelisation epidemiological record reading and validation of the manuscript. All authors read and approved the final manuscript.
Conflict of interest
None declared.
References
- ↑ Wourld Health organization (2016). Global Tuberculosis Report 2016. World Health Organization. pp. 214. ISBN 9789241565394. http://apps.who.int/medicinedocs/en/d/Js23098en/. Retrieved 04 October 2017.
- ↑ 2.0 2.1 Castelnuovo, B.; Kiragga, A.; Afayo, V. et al. (2012). "Implementation of provider-based electronic medical records and improvement of the quality of data in a large HIV program in Sub-Saharan Africa". PLoS One 7 (12): e51631. doi:10.1371/journal.pone.0051631. PMC PMC3524185. PMID 23284728. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3524185.
- ↑ 3.0 3.1 3.2 Rhoads, D.D.; Sintchenko, V.; Rauch, C.A. et al. (2014). "Clinical microbiology informatics". Clinical Microbiology Reviews 27 (4): 1025-47. doi:10.1128/CMR.00049-14. PMC PMC4187636. PMID 25278581. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187636.
- ↑ 4.0 4.1 El-Kareh, R.; Roy, C.; Williams, D.H. et al. (2012). "Impact of automated alerts on follow-up of post-discharge microbiology results: a cluster randomized controlled trial". Journal of General Internal Medicine 27 (10): 1243-50. doi:10.1007/s11606-012-1986-8. PMC PMC3445692. PMID 22278302. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3445692.
- ↑ Ordre National Des Medecins de Côte d’Ivoire (2017). "Plan Strategique de Cybersante". http://doczz.fr/doc/3118472/pdncs-ci---order-national-des-des-decins-de-ivoire-divoire. Retrieved 15 January 2018.
- ↑ Matheron, J.P. (2003). Comprendre Merise: Outils conceptuels et organisationnels (10th ed.). Eyrolles. ISBN 9782212075021.
- ↑ "MySQL Workbench". Oracle Corporation. https://www.mysql.com/fr/products/workbench/. Retrieved 04 October 2017.
- ↑ "Apache Struts". Apache Software Foundation. https://struts.apache.org/. Retrieved 04 October 2017.
- ↑ "Hibernate". Red Hat, Inc. 4 May 2018. http://hibernate.org/.
- ↑ "JasperReports Library". TIBCO Software, Inc. https://community.jaspersoft.com/project/jasperreports-library. Retrieved 16 January 2018.
- ↑ "Apache POI". Apache Software Foundation.
- ↑ Oracle Corporation. "GlassFish: The Open Source Java EE Reference Implementation". Github. https://javaee.github.io/glassfish. Retrieved 14 January 2019.
- ↑ "Date and time input types". Can I use. Patreon. https://caniuse.com/#feat=input-datetime. Retrieved 14 January 2019.
- ↑ Demeuse, M. (2008). "Chapter 5.3 Echelles de Likert ou méthode des classements additionnés" (PDF). Introduction aux théories et aux méthodes de la mesure en sciences psychologiques et en sciences de l'éducation. pp. 213–18. http://iredu.u-bourgogne.fr/images/stories/Documents/Cours_disponibles/Demeuse/Cours/p5.3.pdf. Retrieved 04 January 2018.
- ↑ Cline, G.B.; Luiz, J.M. (2013). "Information technology systems in public sector health facilities in developing countries: the case of South Africa". BMC Medical Informatics and Decision Making 13: 13. doi:10.1186/1472-6947-13-13. PMC PMC3570341. PMID 23347433. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3570341.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. We also added PMCID and DOI when they were missing from the original reference. Under "Architecture and system features," the original article had a poorly punctuated and confusing sentence about browser support for date and time input types; it has been updated to be more correct, with a citation added. The URL to GlassFish was updated to show the 4.x versions. The original had reference 13 in the References section, but it was never referenced in-line; it was omitted in this version.