Difference between revisions of "Main Page/Featured article of the week/2022"
Shawndouglas (talk | contribs) (Added last week's article of the week) |
Shawndouglas (talk | contribs) (Added last week's article of the week) |
||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 31–February 06:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 07–13:</h2> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Xie BMCBioinfo21 22.png|240px]]</div> | |||
'''"[[Journal:Popularity and performance of bioinformatics software: The case of gene set analysis|Popularity and performance of bioinformatics software: The case of gene set analysis]]"''' | |||
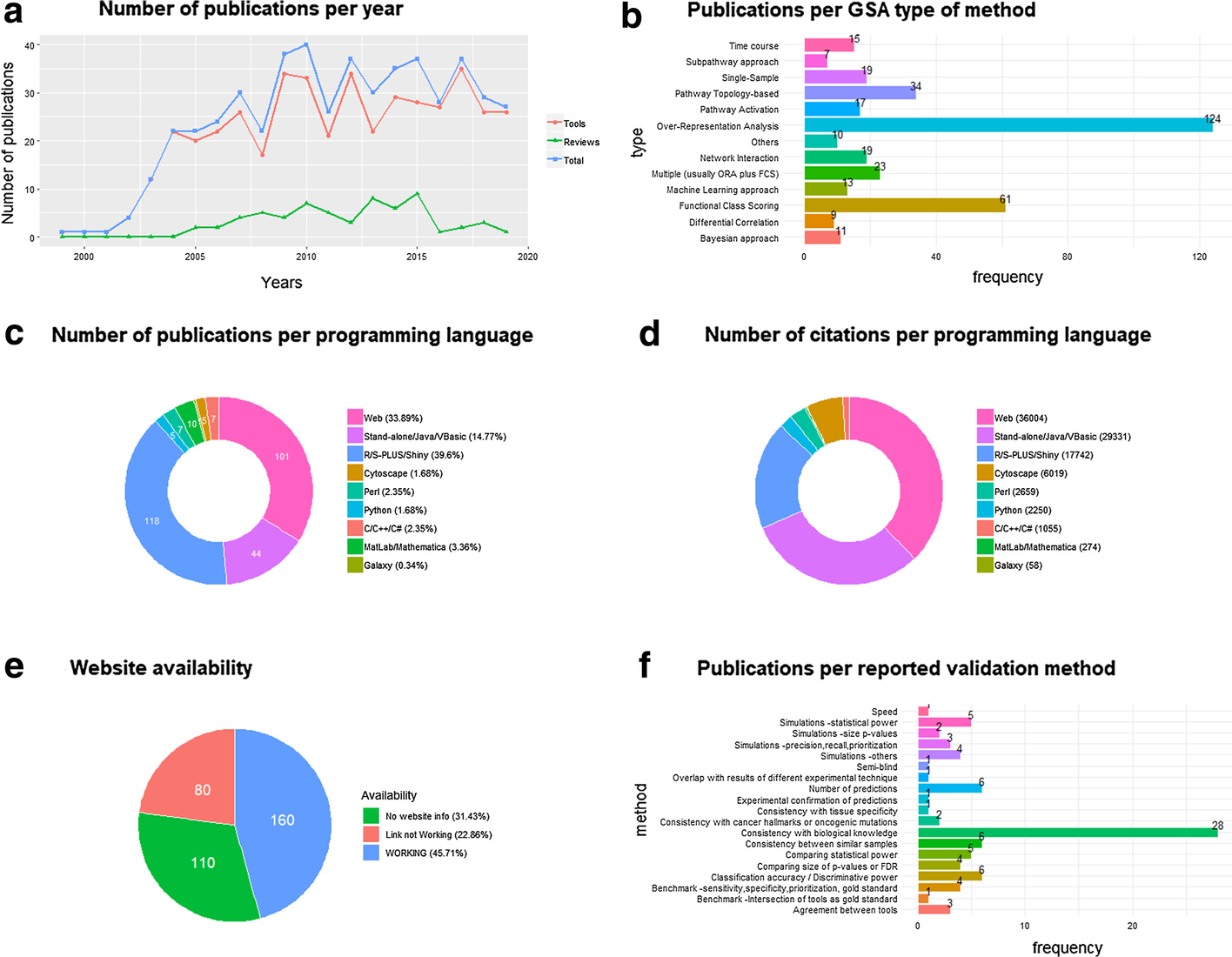
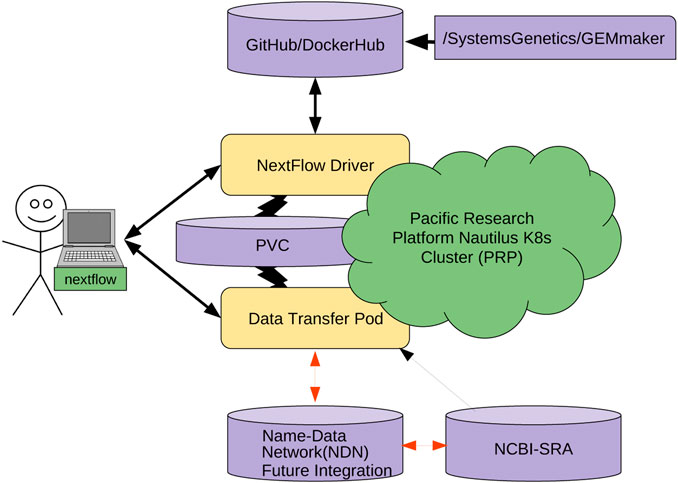
Gene set analysis (GSA) is arguably the method of choice for the functional interpretation of omics results. This work explores the popularity and the performance of all the GSA methodologies and software published during the 20 years since its inception. "Popularity" is estimated according to each paper's citation counts, while "performance" is based on a comprehensive evaluation of the validation strategies used by papers in the field, as well as the consolidated results from the existing benchmark studies. Regarding popularity, data is collected into an online open database ("GSARefDB") which allows browsing bibliographic and method-descriptive [[information]] from 503 GSA paper references; regarding performance, we introduce a repository of [[Jupyter Notebook]] [[workflow]]s and Shiny apps for automated benchmarking of GSA methods (“GSA-BenchmarKING”). After comparing popularity versus performance, results show discrepancies between the most popular and the best performing GSA methods. ('''[[Journal:Popularity and performance of bioinformatics software: The case of gene set analysis|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 31–February 06:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Chong ITMWebConf21 36.png|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Chong ITMWebConf21 36.png|240px]]</div> | ||
'''"[[Journal:Privacy-preserving healthcare informatics: A review|Privacy-preserving healthcare informatics: A review]]"''' | '''"[[Journal:Privacy-preserving healthcare informatics: A review|Privacy-preserving healthcare informatics: A review]]"''' | ||
Revision as of 17:38, 14 February 2022
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 |
Featured article of the week archive - 2022
Welcome to the LIMSwiki 2022 archive for the Featured Article of the Week.
Featured article of the week: February 07–13:"Popularity and performance of bioinformatics software: The case of gene set analysis" Gene set analysis (GSA) is arguably the method of choice for the functional interpretation of omics results. This work explores the popularity and the performance of all the GSA methodologies and software published during the 20 years since its inception. "Popularity" is estimated according to each paper's citation counts, while "performance" is based on a comprehensive evaluation of the validation strategies used by papers in the field, as well as the consolidated results from the existing benchmark studies. Regarding popularity, data is collected into an online open database ("GSARefDB") which allows browsing bibliographic and method-descriptive information from 503 GSA paper references; regarding performance, we introduce a repository of Jupyter Notebook workflows and Shiny apps for automated benchmarking of GSA methods (“GSA-BenchmarKING”). After comparing popularity versus performance, results show discrepancies between the most popular and the best performing GSA methods. (Full article...)
|