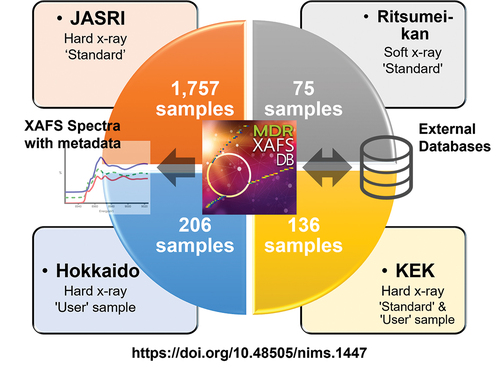
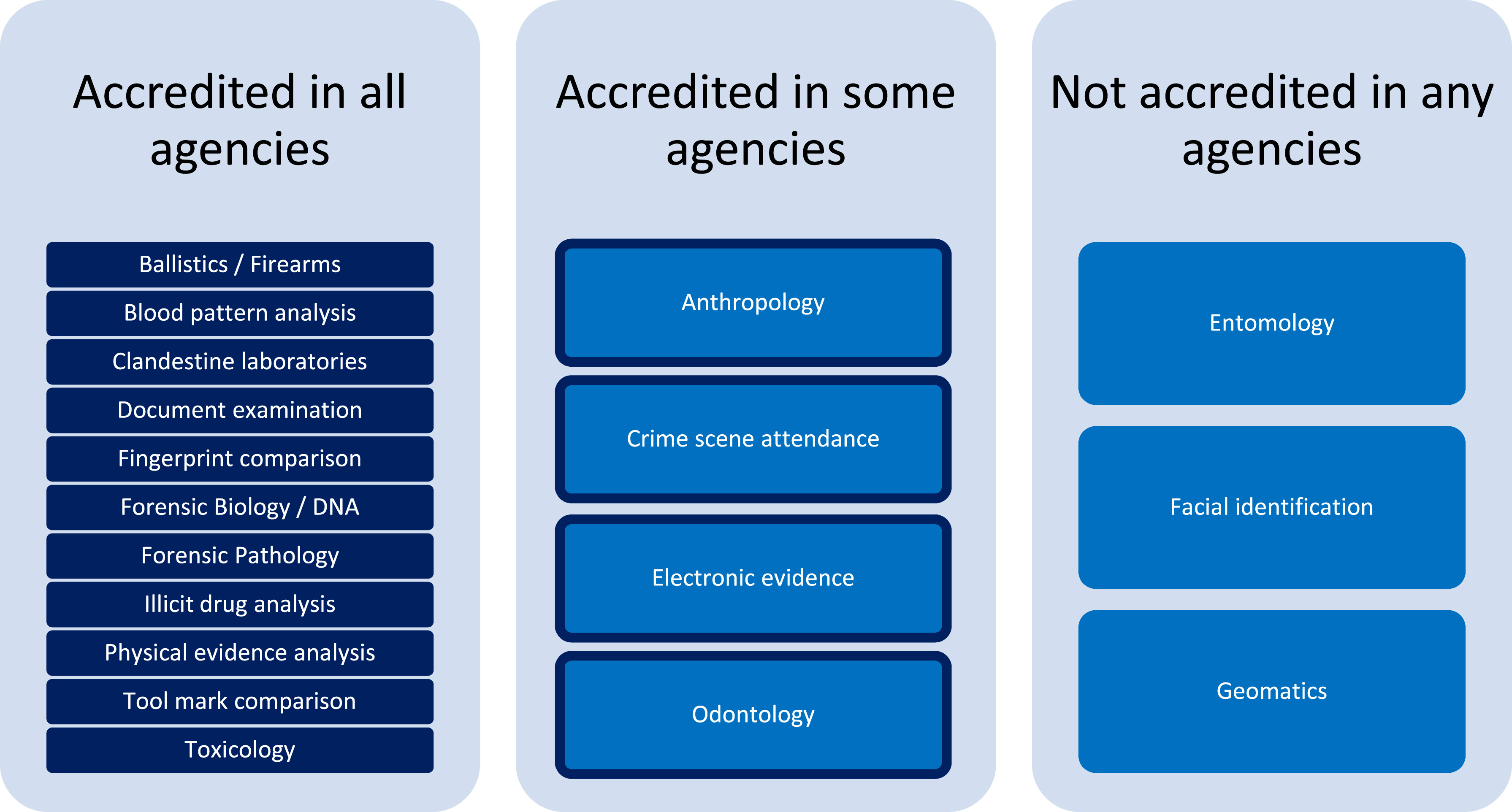
Difference between revisions of "Main Page/Featured article of the week/2024"
Shawndouglas (talk | contribs) (Added last week's article of the week) |
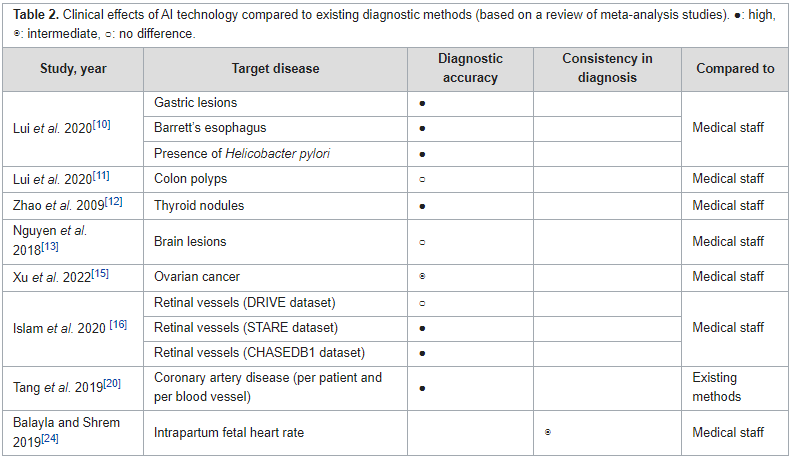
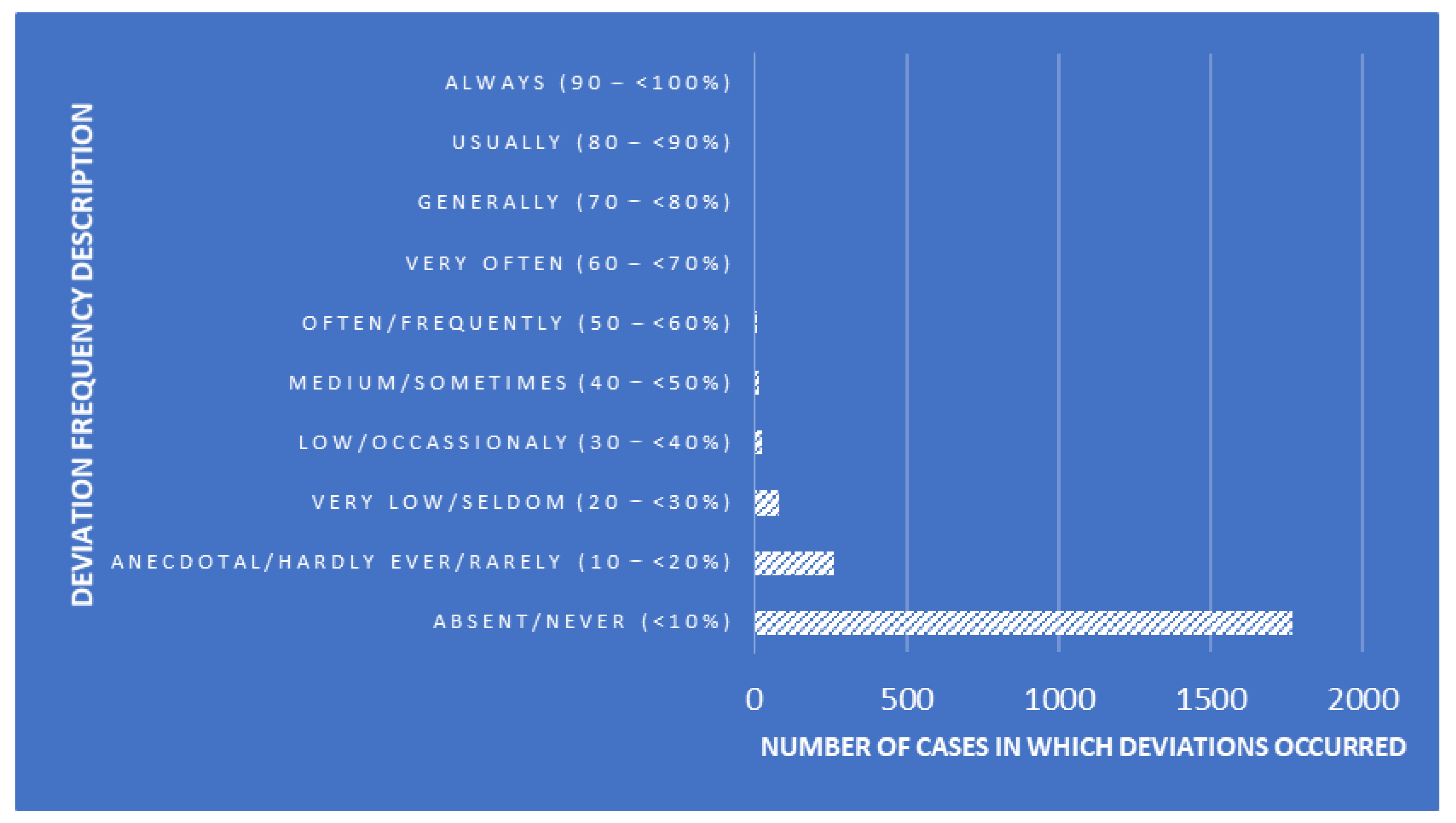
Shawndouglas (talk | contribs) (Added last week's article of the week) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 29–May 05:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 10–16:</h2> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Berezin PLoSCompBio23 19-12.png|240px]]</div> | |||
'''"[[Journal:Ten simple rules for managing laboratory information|Ten simple rules for managing laboratory information]]"''' | |||
[[Information]] is the cornerstone of [[research]], from experimental data/[[metadata]] and computational processes to complex inventories of reagents and equipment. These 10 simple rules discuss best practices for leveraging [[laboratory information management system]]s (LIMS) to transform this large information load into useful scientific findings. The development of [[mathematical model]]s that can predict the properties of biological systems is the holy grail of [[computational biology]]. Such models can be used to test biological hypotheses, guide the development of biomanufactured products, engineer new systems meeting user-defined specifications, and much more ... ('''[[Journal:Ten simple rules for managing laboratory information|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 03–09:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Signoroni NatComm23 14.png|240px]]</div> | |||
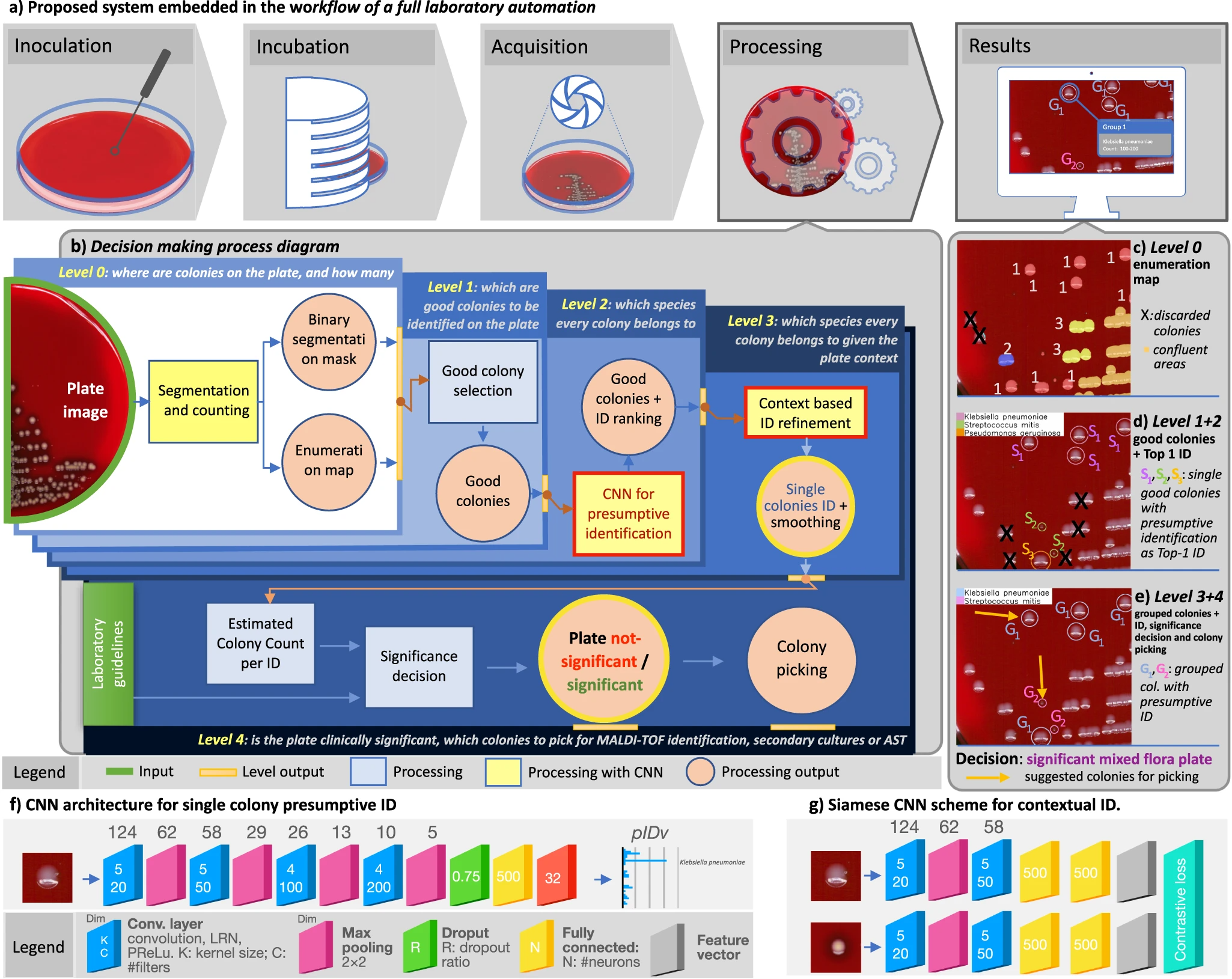
'''"[[Journal:Hierarchical AI enables global interpretation of culture plates in the era of digital microbiology|Hierarchical AI enables global interpretation of culture plates in the era of digital microbiology]]"''' | |||
Full [[laboratory automation]] is revolutionizing work habits in an increasing number of clinical [[microbiology]] facilities worldwide, generating huge streams of [[Imaging|digital images]] for interpretation. Contextually, [[deep learning]] (DL) architectures are leading to paradigm shifts in the way computers can assist with difficult visual interpretation tasks in several domains. At the crossroads of these epochal trends, we present a system able to tackle a core task in clinical microbiology, namely the global interpretation of diagnostic [[Bacteria|bacterial]] [[Cell culture|culture]] plates, including presumptive [[pathogen]] identification. This is achieved by decomposing the problem into a hierarchy of complex subtasks and addressing them with a multi-network architecture we call DeepColony ... ('''[[Journal:Hierarchical AI enables global interpretation of culture plates in the era of digital microbiology|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 27–June 02:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Čartolovni DigitalHealth2023 9.jpeg|240px]]</div> | |||
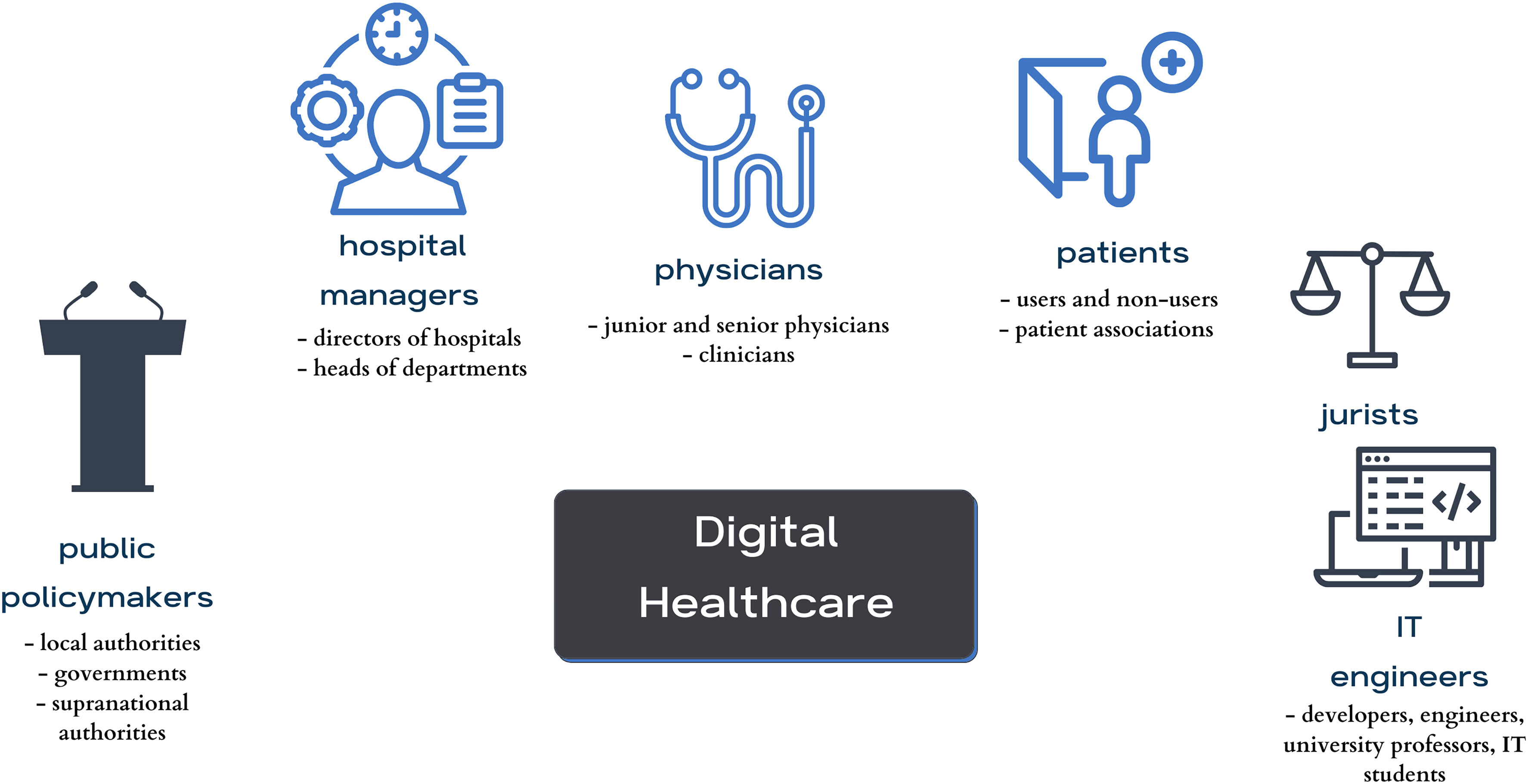
'''"[[Journal:Critical analysis of the impact of AI on the patient–physician relationship: A multi-stakeholder qualitative study|Critical analysis of the impact of AI on the patient–physician relationship: A multi-stakeholder qualitative study]]"''' | |||
This qualitative study aims to present the aspirations, expectations, and critical analysis of the potential for [[artificial intelligence]] (AI) to transform the patient–physician relationship, according to multi-stakeholder insight. This study was conducted from June to December 2021, using an anticipatory ethics approach and sociology of expectations as the theoretical frameworks. It focused mainly on three groups of stakeholders, namely physicians (''n'' = 12), patients (''n'' = 15), and healthcare managers (''n'' = 11), all of whom are directly related to the adoption of AI in medicine (''n'' = 38). In this study, interviews were conducted with 40% of the patients in the sample (15/38), as well as 31% of the physicians (12/38) and 29% of health managers in the sample (11/38) ... ('''[[Journal:Critical analysis of the impact of AI on the patient–physician relationship: A multi-stakeholder qualitative study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 20–26:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Niszczota EconBusRev23 9-2.png|240px]]</div> | |||
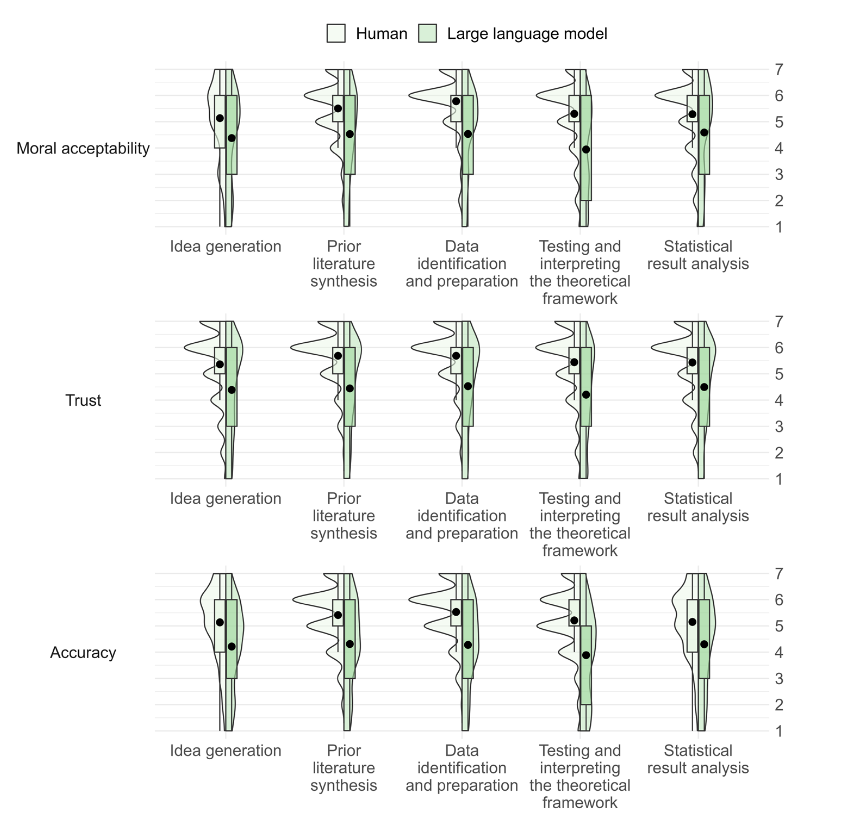
'''"[[Journal:Judgements of research co-created by generative AI: Experimental evidence|Judgements of research co-created by generative AI: Experimental evidence]]"''' | |||
The introduction of [[ChatGPT]] has fuelled a public debate on the appropriateness of using generative [[artificial intelligence]] (AI) ([[large language model]]s or LLMs) in work, including a debate on how they might be used (and abused) by researchers. In the current work, we test whether delegating parts of the research process to LLMs leads people to distrust researchers and devalues their scientific work. Participants (''N'' = 402) considered a researcher who delegates elements of the research process to a PhD student or LLM and rated three aspects of such delegation. Firstly, they rated whether it is morally appropriate to do so. Secondly, they judged whether—after deciding to delegate the research process—they would trust the scientist (who decided to delegate) to oversee future projects ... ('''[[Journal:Judgements of research co-created by generative AI: Experimental evidence|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 13–19:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Bispo-Silva Geosciences23 13-11.png|240px]]</div> | |||
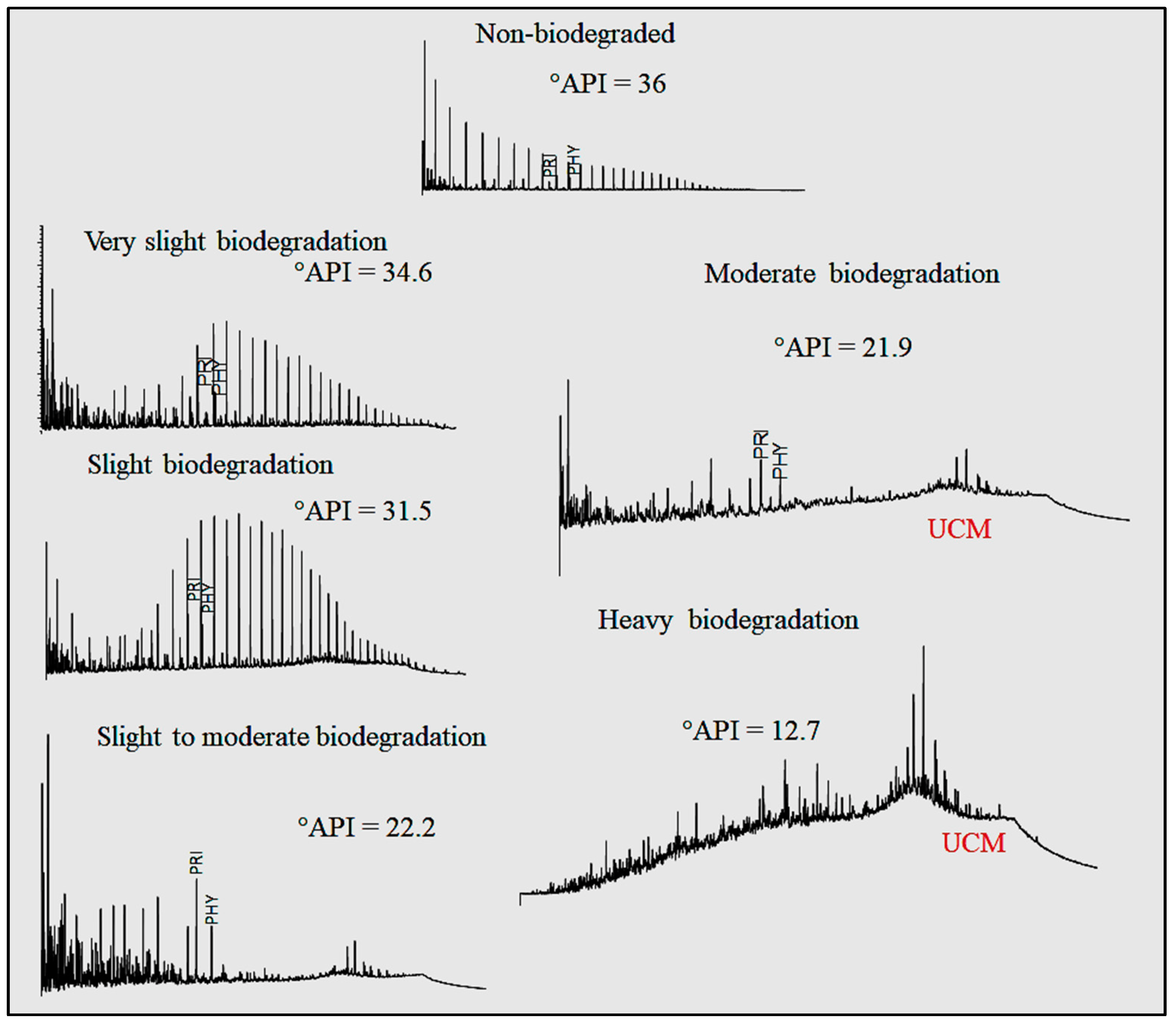
'''"[[Journal:Geochemical biodegraded oil classification using a machine learning approach|Geochemical biodegraded oil classification using a machine learning approach]]"''' | |||
[[Chromatography|Chromatographic]] oil analysis is an important step for the identification of biodegraded petroleum via peak visualization and interpretation of phenomena that explain the oil geochemistry. However, analyses of chromatogram components by geochemists are comparative, visual, and consequently slow. This article aims to improve the chromatogram analysis process performed during geochemical interpretation by proposing the use of [[convolutional neural network]]s (CNN), which are deep learning techniques widely used by big tech companies. Two hundred and twenty-one (221) chromatographic oil images from different worldwide basins (Brazil, USA, Portugal, Angola, and Venezuela) were used. The [[open-source software]] Orange Data Mining was used to process images by CNN. The CNN algorithm extracts, pixel by pixel, recurring features from the images through convolutional operations ... ('''[[Journal:Geochemical biodegraded oil classification using a machine learning approach|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 06–12:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Mishra JofNepMedAss23 61-258.png|220px]]</div> | |||
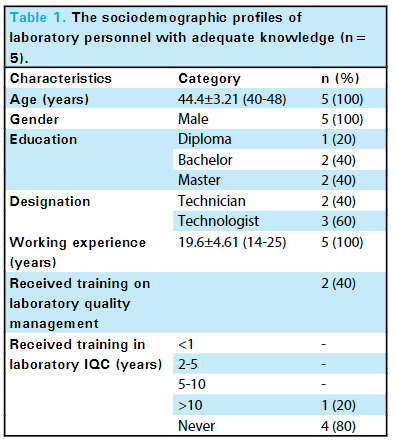
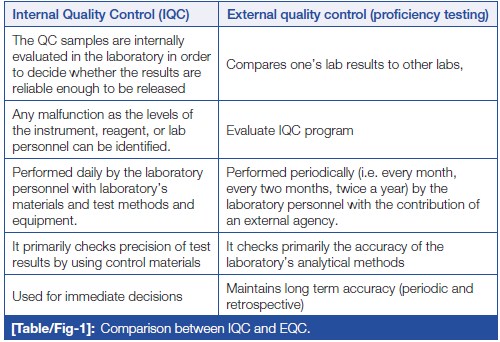
'''"[[Journal:Knowledge of internal quality control for laboratory tests among laboratory personnel working in a biochemistry department of a tertiary care center: A descriptive cross-sectional study|Knowledge of internal quality control for laboratory tests among laboratory personnel working in a biochemistry department of a tertiary care center: A descriptive cross-sectional study]]"''' | |||
The [[clinical laboratory]] holds a central position in patient care, and as such, ensuring accurate [[laboratory]] test results is a necessity. Internal [[quality control]] (QC) ensures day-to-day laboratory consistency. However, unless practiced, the success of laboratory [[quality management system]]s (QMSs) cannot be achieved. This depends on the efforts and commitment of laboratory personnel for its implementation. Hence, the aim of this study was to find out the knowledge of internal QC for laboratory tests among laboratory personnel working in the Department of Biochemistry, B.P. Koirala Institute of Health Sciences (BPKIHS), a tertiary care center ... ('''[[Journal:Knowledge of internal quality control for laboratory tests among laboratory personnel working in a biochemistry department of a tertiary care center: A descriptive cross-sectional study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 29–May 05:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Karaattuthazhathu NatJLabMed23 12-2.png|260px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Karaattuthazhathu NatJLabMed23 12-2.png|260px]]</div> | ||
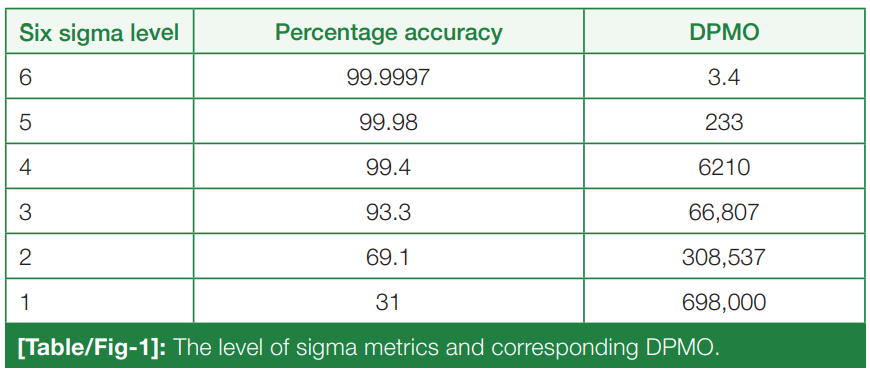
'''"[[Journal:Sigma metrics as a valuable tool for effective analytical performance and quality control planning in the clinical laboratory: A retrospective study|Sigma metrics as a valuable tool for effective analytical performance and quality control planning in the clinical laboratory: A retrospective study]]"''' | '''"[[Journal:Sigma metrics as a valuable tool for effective analytical performance and quality control planning in the clinical laboratory: A retrospective study|Sigma metrics as a valuable tool for effective analytical performance and quality control planning in the clinical laboratory: A retrospective study]]"''' | ||
Latest revision as of 15:03, 17 June 2024
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 - 2023 - 2024 |
Featured article of the week archive - 2024
Welcome to the LIMSwiki 2024 archive for the Featured Article of the Week.
Featured article of the week: June 10–16:"Ten simple rules for managing laboratory information" Information is the cornerstone of research, from experimental data/metadata and computational processes to complex inventories of reagents and equipment. These 10 simple rules discuss best practices for leveraging laboratory information management systems (LIMS) to transform this large information load into useful scientific findings. The development of mathematical models that can predict the properties of biological systems is the holy grail of computational biology. Such models can be used to test biological hypotheses, guide the development of biomanufactured products, engineer new systems meeting user-defined specifications, and much more ... (Full article...)
|