Difference between revisions of "Main Page/Featured article of the week/2024"
Shawndouglas (talk | contribs) (Added last week's article of the week) |
Shawndouglas (talk | contribs) (Added last week's article of the week) |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 15–21:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 10–16:</h2> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Berezin PLoSCompBio23 19-12.png|240px]]</div> | |||
'''"[[Journal:Ten simple rules for managing laboratory information|Ten simple rules for managing laboratory information]]"''' | |||
[[Information]] is the cornerstone of [[research]], from experimental data/[[metadata]] and computational processes to complex inventories of reagents and equipment. These 10 simple rules discuss best practices for leveraging [[laboratory information management system]]s (LIMS) to transform this large information load into useful scientific findings. The development of [[mathematical model]]s that can predict the properties of biological systems is the holy grail of [[computational biology]]. Such models can be used to test biological hypotheses, guide the development of biomanufactured products, engineer new systems meeting user-defined specifications, and much more ... ('''[[Journal:Ten simple rules for managing laboratory information|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: June 03–09:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Signoroni NatComm23 14.png|240px]]</div> | |||
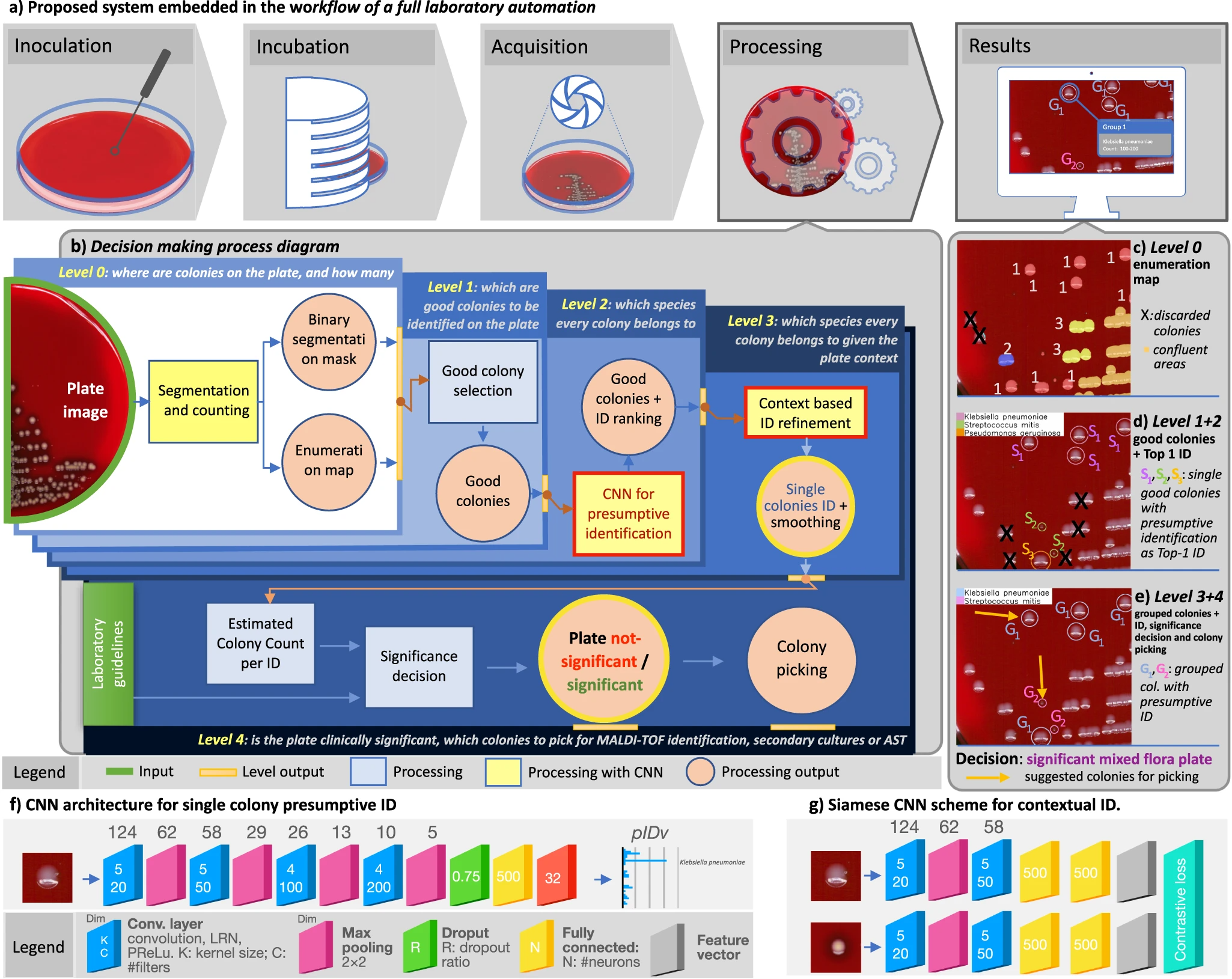
'''"[[Journal:Hierarchical AI enables global interpretation of culture plates in the era of digital microbiology|Hierarchical AI enables global interpretation of culture plates in the era of digital microbiology]]"''' | |||
Full [[laboratory automation]] is revolutionizing work habits in an increasing number of clinical [[microbiology]] facilities worldwide, generating huge streams of [[Imaging|digital images]] for interpretation. Contextually, [[deep learning]] (DL) architectures are leading to paradigm shifts in the way computers can assist with difficult visual interpretation tasks in several domains. At the crossroads of these epochal trends, we present a system able to tackle a core task in clinical microbiology, namely the global interpretation of diagnostic [[Bacteria|bacterial]] [[Cell culture|culture]] plates, including presumptive [[pathogen]] identification. This is achieved by decomposing the problem into a hierarchy of complex subtasks and addressing them with a multi-network architecture we call DeepColony ... ('''[[Journal:Hierarchical AI enables global interpretation of culture plates in the era of digital microbiology|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 27–June 02:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Čartolovni DigitalHealth2023 9.jpeg|240px]]</div> | |||
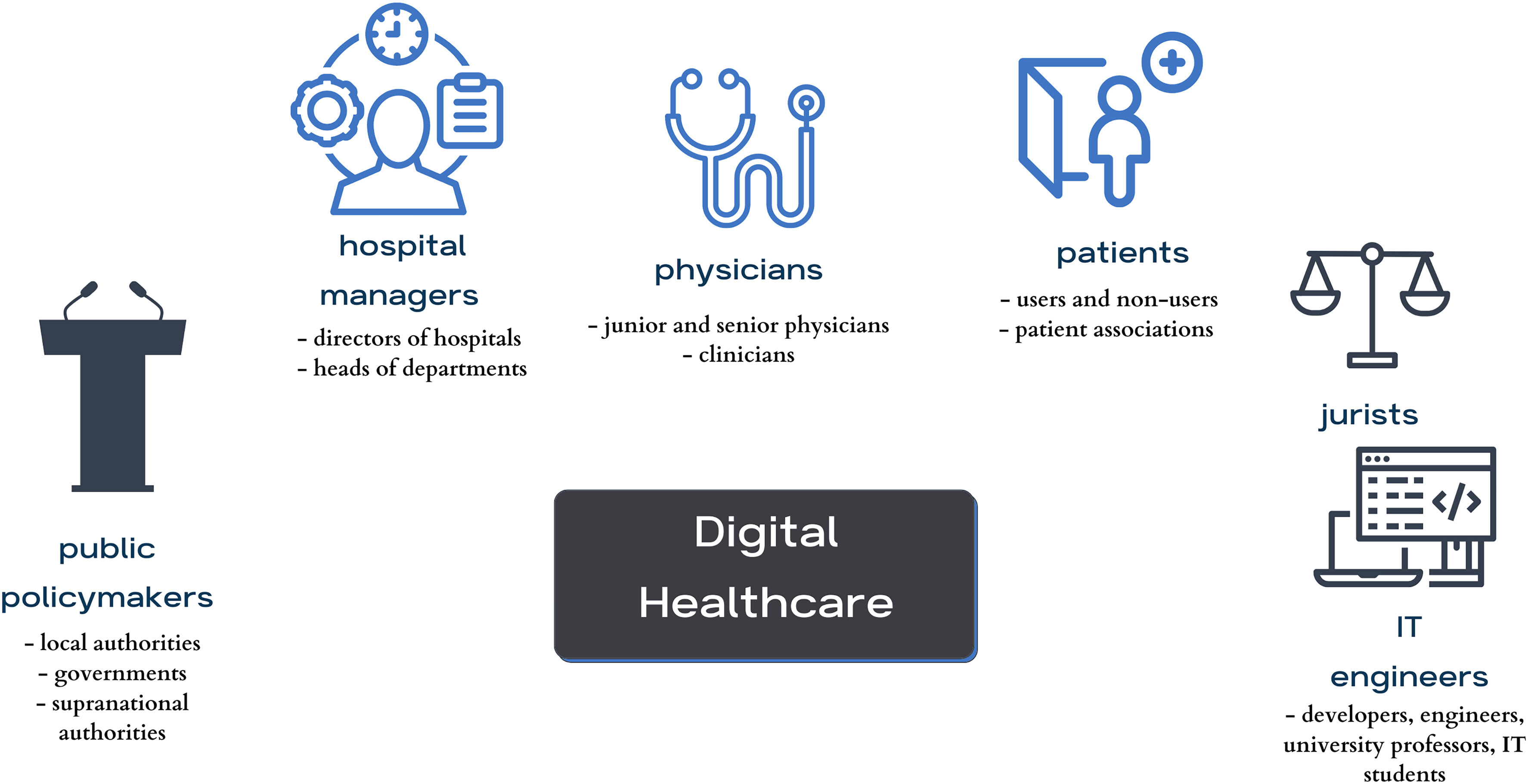
'''"[[Journal:Critical analysis of the impact of AI on the patient–physician relationship: A multi-stakeholder qualitative study|Critical analysis of the impact of AI on the patient–physician relationship: A multi-stakeholder qualitative study]]"''' | |||
This qualitative study aims to present the aspirations, expectations, and critical analysis of the potential for [[artificial intelligence]] (AI) to transform the patient–physician relationship, according to multi-stakeholder insight. This study was conducted from June to December 2021, using an anticipatory ethics approach and sociology of expectations as the theoretical frameworks. It focused mainly on three groups of stakeholders, namely physicians (''n'' = 12), patients (''n'' = 15), and healthcare managers (''n'' = 11), all of whom are directly related to the adoption of AI in medicine (''n'' = 38). In this study, interviews were conducted with 40% of the patients in the sample (15/38), as well as 31% of the physicians (12/38) and 29% of health managers in the sample (11/38) ... ('''[[Journal:Critical analysis of the impact of AI on the patient–physician relationship: A multi-stakeholder qualitative study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 20–26:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Niszczota EconBusRev23 9-2.png|240px]]</div> | |||
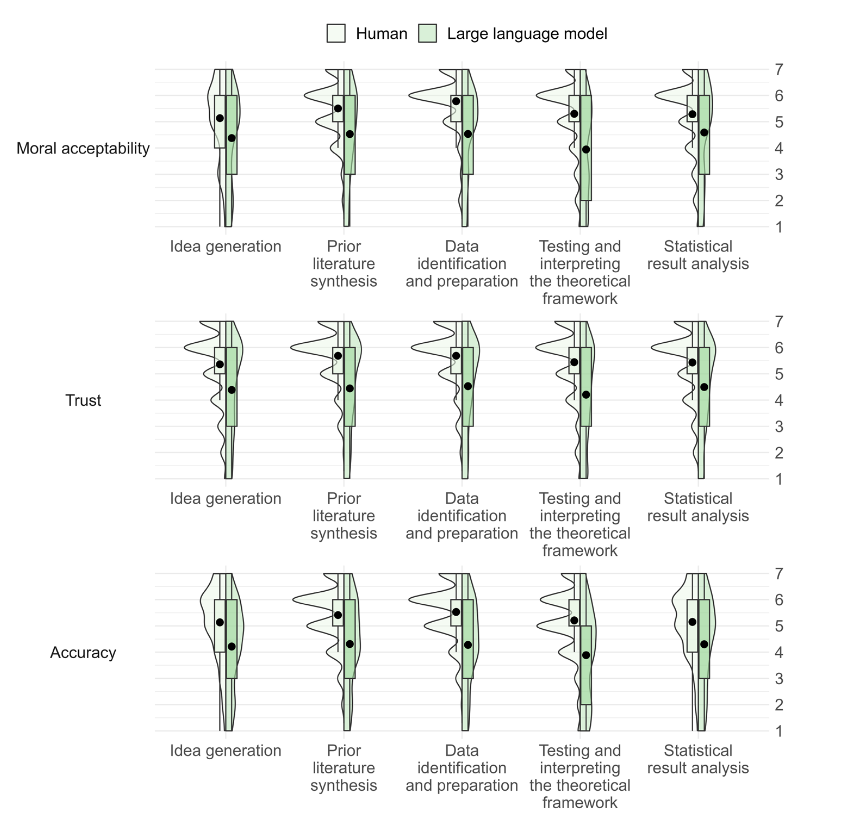
'''"[[Journal:Judgements of research co-created by generative AI: Experimental evidence|Judgements of research co-created by generative AI: Experimental evidence]]"''' | |||
The introduction of [[ChatGPT]] has fuelled a public debate on the appropriateness of using generative [[artificial intelligence]] (AI) ([[large language model]]s or LLMs) in work, including a debate on how they might be used (and abused) by researchers. In the current work, we test whether delegating parts of the research process to LLMs leads people to distrust researchers and devalues their scientific work. Participants (''N'' = 402) considered a researcher who delegates elements of the research process to a PhD student or LLM and rated three aspects of such delegation. Firstly, they rated whether it is morally appropriate to do so. Secondly, they judged whether—after deciding to delegate the research process—they would trust the scientist (who decided to delegate) to oversee future projects ... ('''[[Journal:Judgements of research co-created by generative AI: Experimental evidence|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 13–19:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Bispo-Silva Geosciences23 13-11.png|240px]]</div> | |||
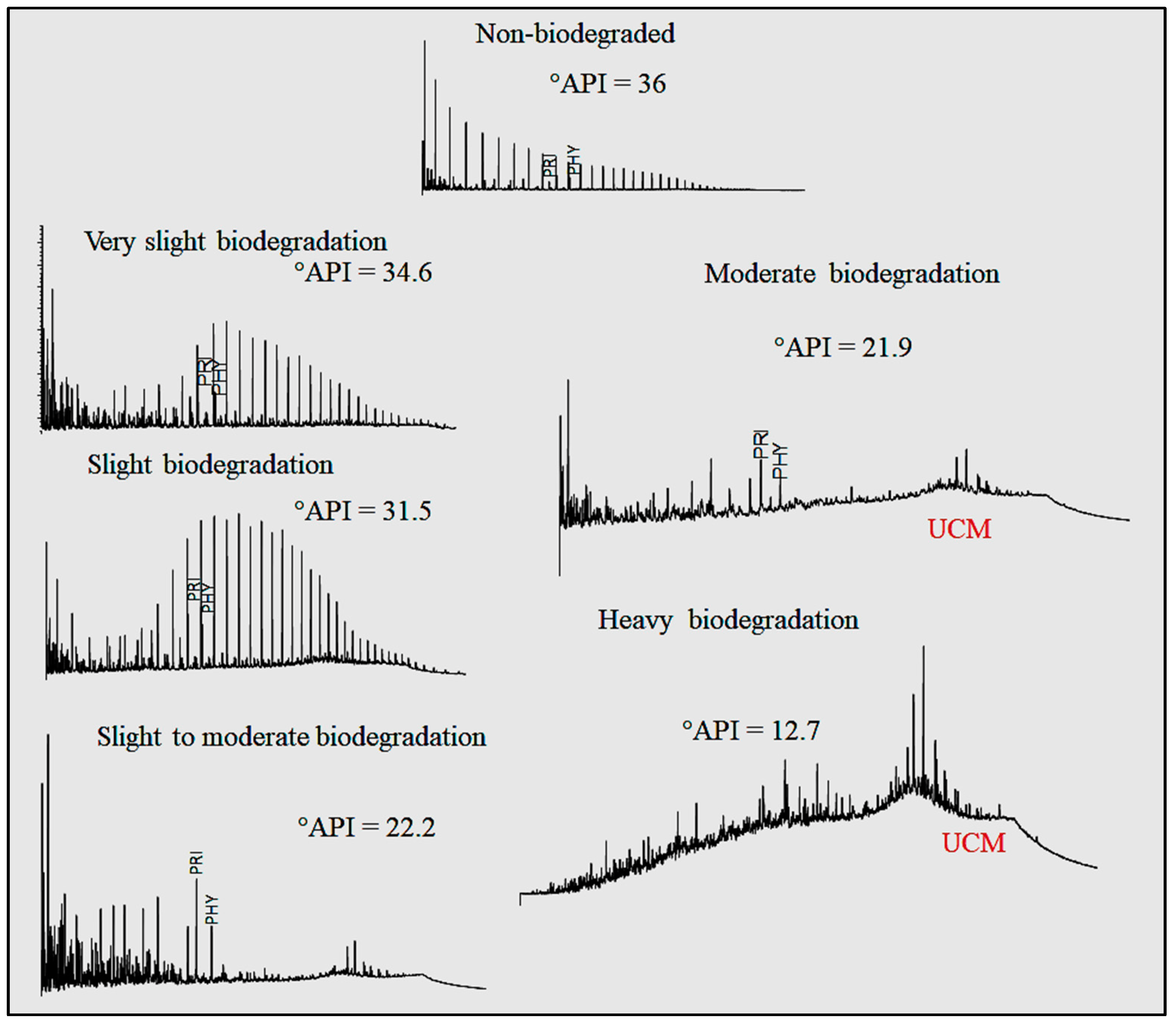
'''"[[Journal:Geochemical biodegraded oil classification using a machine learning approach|Geochemical biodegraded oil classification using a machine learning approach]]"''' | |||
[[Chromatography|Chromatographic]] oil analysis is an important step for the identification of biodegraded petroleum via peak visualization and interpretation of phenomena that explain the oil geochemistry. However, analyses of chromatogram components by geochemists are comparative, visual, and consequently slow. This article aims to improve the chromatogram analysis process performed during geochemical interpretation by proposing the use of [[convolutional neural network]]s (CNN), which are deep learning techniques widely used by big tech companies. Two hundred and twenty-one (221) chromatographic oil images from different worldwide basins (Brazil, USA, Portugal, Angola, and Venezuela) were used. The [[open-source software]] Orange Data Mining was used to process images by CNN. The CNN algorithm extracts, pixel by pixel, recurring features from the images through convolutional operations ... ('''[[Journal:Geochemical biodegraded oil classification using a machine learning approach|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: May 06–12:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Mishra JofNepMedAss23 61-258.png|220px]]</div> | |||
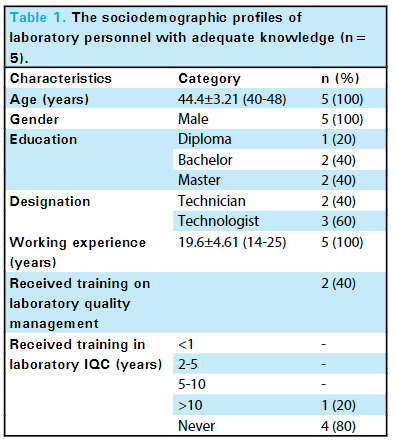
'''"[[Journal:Knowledge of internal quality control for laboratory tests among laboratory personnel working in a biochemistry department of a tertiary care center: A descriptive cross-sectional study|Knowledge of internal quality control for laboratory tests among laboratory personnel working in a biochemistry department of a tertiary care center: A descriptive cross-sectional study]]"''' | |||
The [[clinical laboratory]] holds a central position in patient care, and as such, ensuring accurate [[laboratory]] test results is a necessity. Internal [[quality control]] (QC) ensures day-to-day laboratory consistency. However, unless practiced, the success of laboratory [[quality management system]]s (QMSs) cannot be achieved. This depends on the efforts and commitment of laboratory personnel for its implementation. Hence, the aim of this study was to find out the knowledge of internal QC for laboratory tests among laboratory personnel working in the Department of Biochemistry, B.P. Koirala Institute of Health Sciences (BPKIHS), a tertiary care center ... ('''[[Journal:Knowledge of internal quality control for laboratory tests among laboratory personnel working in a biochemistry department of a tertiary care center: A descriptive cross-sectional study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 29–May 05:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Karaattuthazhathu NatJLabMed23 12-2.png|260px]]</div> | |||
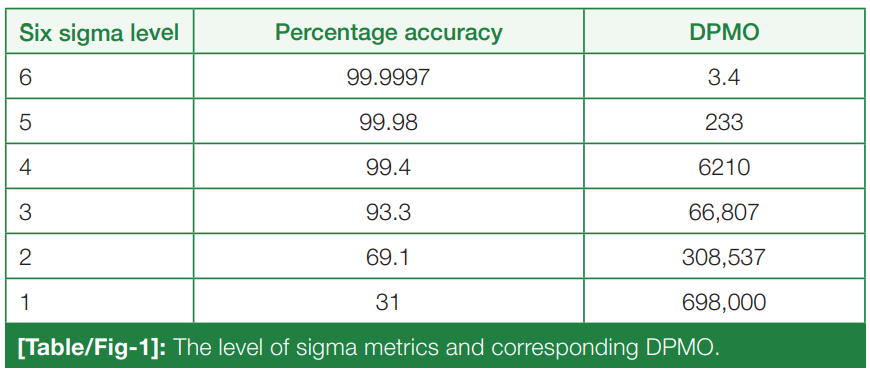
'''"[[Journal:Sigma metrics as a valuable tool for effective analytical performance and quality control planning in the clinical laboratory: A retrospective study|Sigma metrics as a valuable tool for effective analytical performance and quality control planning in the clinical laboratory: A retrospective study]]"''' | |||
For the release of precise and accurate reports of [[Medical test|routine tests]], its necessary to follow a proper [[quality management system]] (QMS) in the [[clinical laboratory]]. As one of the most popular QMS tools for process improvement, Six Sigma techniques and tools have been accepted widely in the [[laboratory]] testing process. Six Sigma gives an objective assessment of analytical methods and instrumentation, measuring the outcome of a process on a scale of 0 to 6. Poor outcomes are measured in terms of defects per million opportunities (DPMO). To do the performance assessment of each clinical laboratory [[analyte]] by Six Sigma analysis and to plan and chart out a better, customized [[quality control]] (QC) plan for each analyte, according to its own sigma value ... ('''[[Journal:Sigma metrics as a valuable tool for effective analytical performance and quality control planning in the clinical laboratory: A retrospective study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 22–28:</h2> | |||
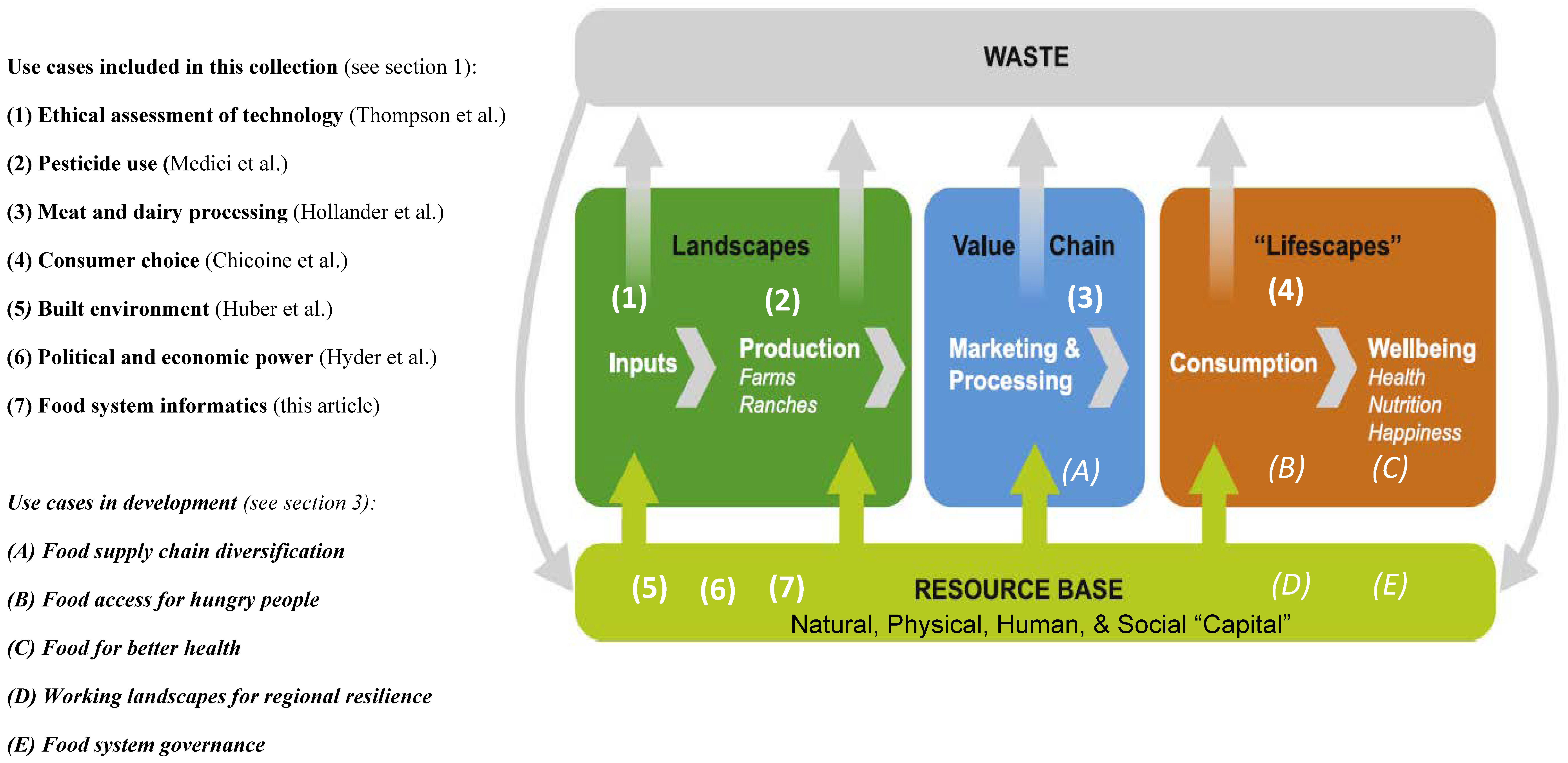
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Tomich Sustain23 15-8.png|260px]]</div> | |||
'''"[[Journal:Why do we need food systems informatics? Introduction to this special collection on smart and connected regional food systems|Why do we need food systems informatics? Introduction to this special collection on smart and connected regional food systems]]"''' | |||
Public interest in where food comes from and how it is produced, processed, and distributed has increased over the last few decades, with even greater focus emerging during the [[COVID-19]] [[pandemic]]. Mounting evidence and experience point to disturbing weaknesses in our food systems’ abilities to support human livelihoods and wellbeing, and alarming long-term trends regarding both the environmental footprint of food systems and mounting vulnerabilities to shocks and stressors. How can we tackle the “wicked problems” embedded in a food system? More specifically, how can convergent research programs be designed and resulting knowledge implemented to increase inclusion, sustainability, and resilience within these complex systems ... ('''[[Journal:Why do we need food systems informatics? Introduction to this special collection on smart and connected regional food systems|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 15–21:</h2> | |||
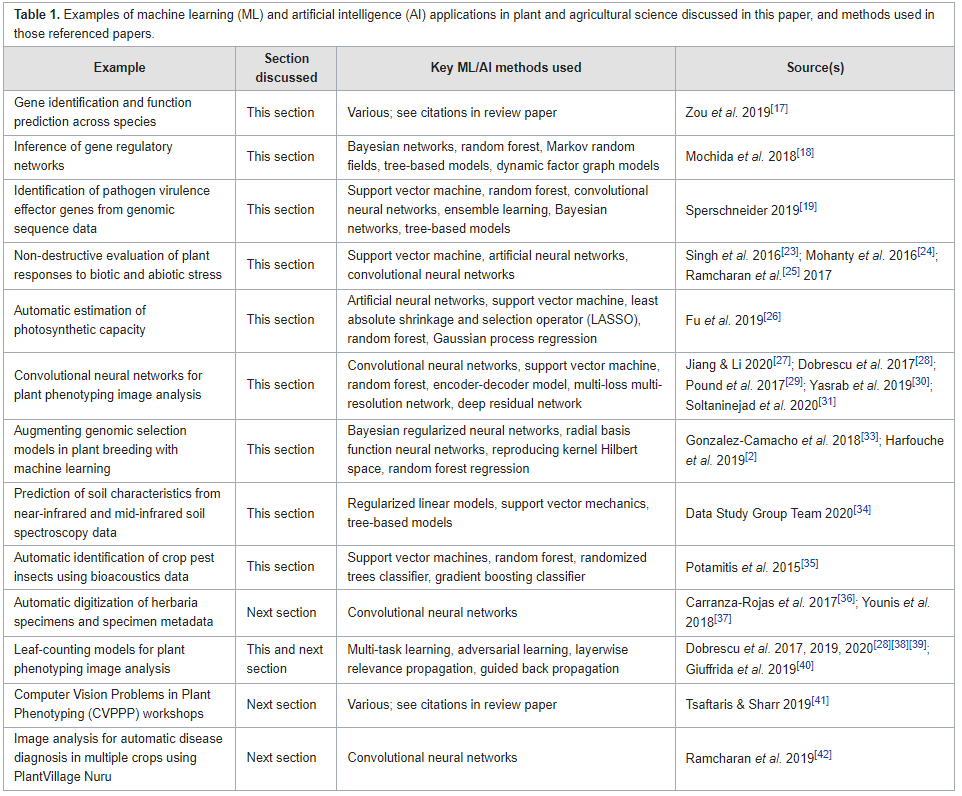
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab1 Williamson F1000Res2023 10.png|240px]]</div> | |||
'''"[[Journal:Data management challenges for artificial intelligence in plant and agricultural research|Data management challenges for artificial intelligence in plant and agricultural research]]"''' | |||
[[Artificial intelligence]] (AI) is increasingly used within plant science, yet it is far from being routinely and effectively implemented in this domain. Particularly relevant to the development of novel food and agricultural technologies is the development of validated, meaningful, and usable ways to integrate, compare, and [[Data visualization|visualize]] large, multi-dimensional datasets from different sources and scientific approaches. After a brief summary of the reasons for the interest in data science and AI within plant science, the paper identifies and discusses eight key challenges in [[Information management|data management]] that must be addressed to further unlock the potential of AI in crop and agronomic research, and particularly the application of [[machine learning]] (ML), which holds much promise for this domain ... ('''[[Journal:Data management challenges for artificial intelligence in plant and agricultural research|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 08–14:</h2> | |||
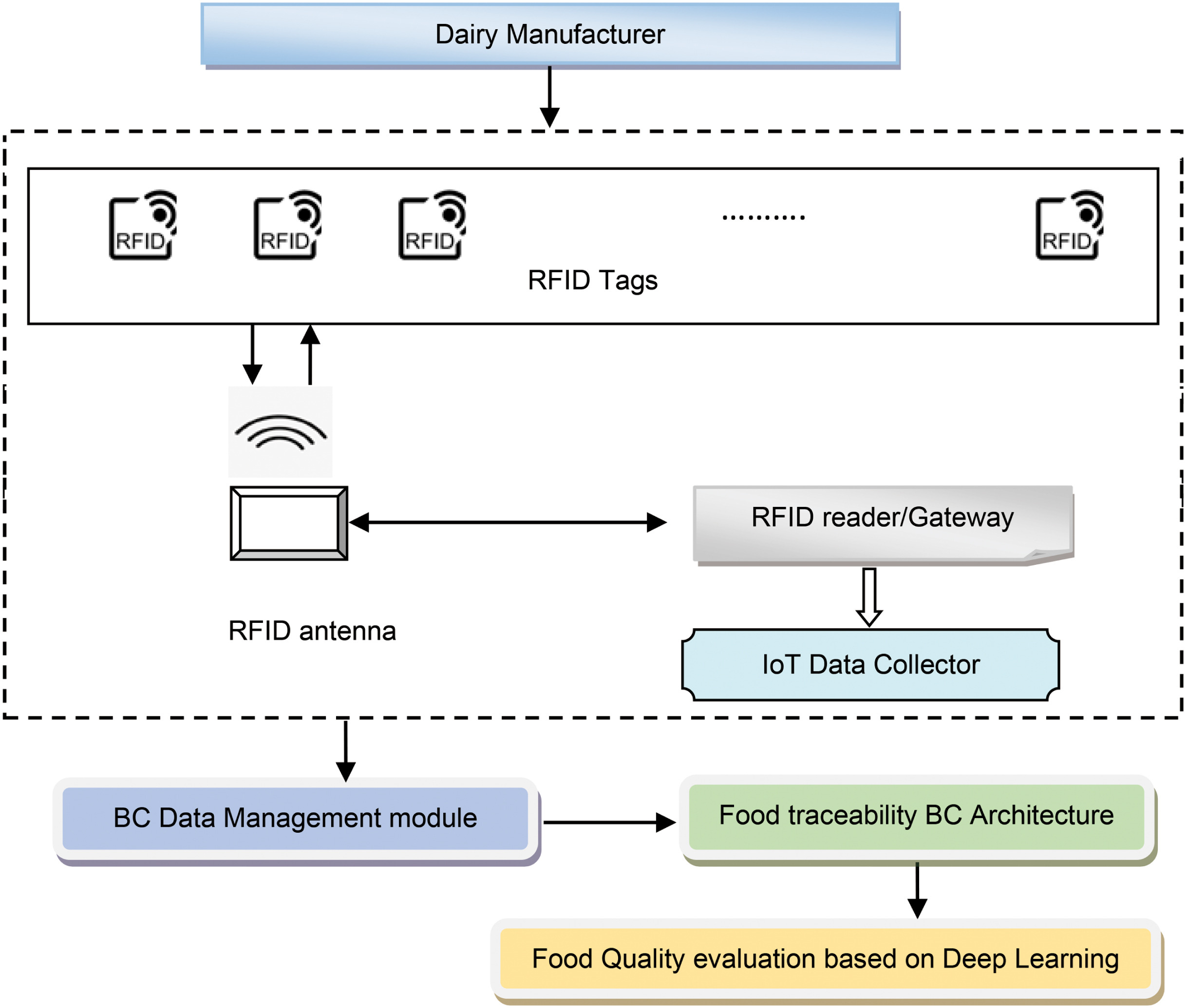
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Manisha HighConComp2023 3-3.jpg|240px]]</div> | |||
'''"[[Journal:A blockchain-driven IoT-based food quality traceability system for dairy products using a deep learning model|A blockchain-driven IoT-based food quality traceability system for dairy products using a deep learning model]]"''' | |||
Food [[traceability]] is a critical factor that can ensure food safety while enhancing the credibility of the manufactured product, thus achieving heightened user satisfaction and loyalty. The perishable food supply chain (PFSC) requires paramount care for ensuring [[Quality (business)|quality]] owing to the limited product life. The PFSC comprises of multiple organizations with varied interests and is more likely to be hesitant in sharing the traceability details among one another owing to a lack of trust, which can be overcome by using blockchain. In this research, an efficient scheme using a blockchain-enabled deep [[wikipedia:Residual neural network|residual network]] (BC-DRN) is developed to provide food traceability for dairy products. Here, food traceability is determined by using various modular tools ... ('''[[Journal:A blockchain-driven IoT-based food quality traceability system for dairy products using a deep learning model|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: April 01–07:</h2> | |||
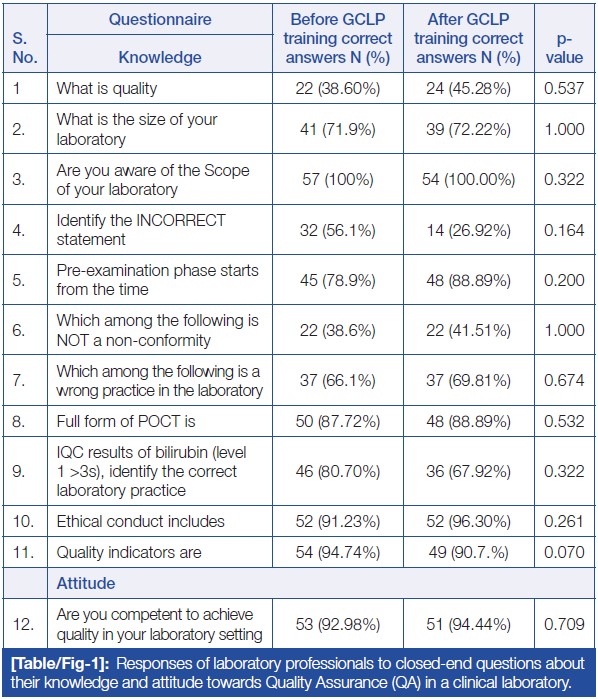
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Patel JofClinDiagRes2023 17-9.jpg|140px]]</div> | |||
'''"[[Journal:Effect of good clinical laboratory practices (GCLP) quality training on knowledge, attitude, and practice among laboratory professionals: Quasi-experimental study|Effect of good clinical laboratory practices (GCLP) quality training on knowledge, attitude, and practice among laboratory professionals: Quasi-experimental study]]"''' | |||
Good clinical laboratory practices (GCLP) play a vital role in early and accurate diagnosis, providing high-quality data and timely [[Sample (material)|sample]] processing. Adhering to a robust [[quality management system]] (QMS) that complies with GCLP standards is crucial for [[laboratory]] personnel in a [[clinical laboratory]] to deliver outstanding healthcare services and reliable, reproducible reports. [The aim of this study is to] assess the knowledge, attitude, and practice (KAP) of laboratory professionals towards [[Quality (business)|quality]] in the laboratory through GCLP training... ('''[[Journal:Effect of good clinical laboratory practices (GCLP) quality training on knowledge, attitude, and practice among laboratory professionals: Quasi-experimental study|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 25–31:</h2> | |||
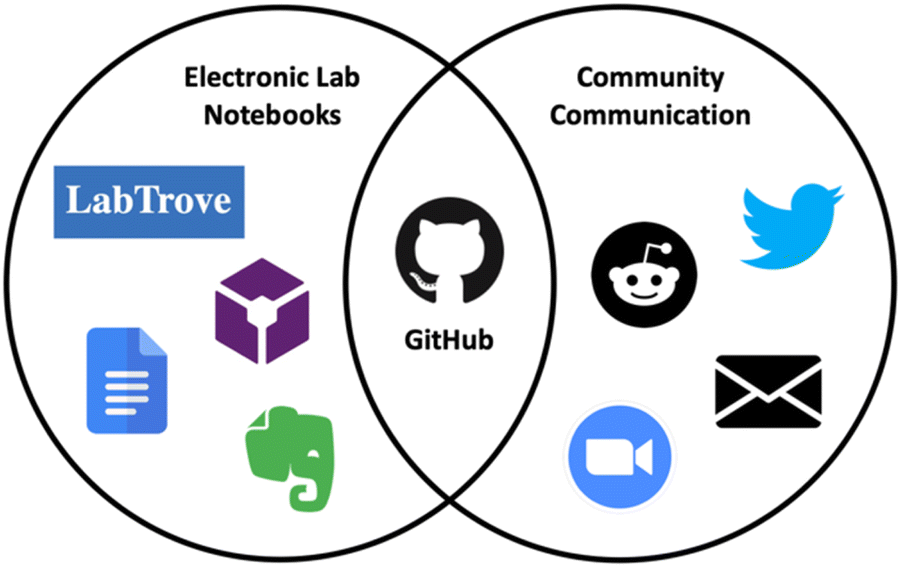
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Scroggie DigDisc2023 2.gif|240px]]</div> | |||
'''"[[Journal:GitHub as an open electronic laboratory notebook for real-time sharing of knowledge and collaboration|GitHub as an open electronic laboratory notebook for real-time sharing of knowledge and collaboration]]"''' | |||
[[Electronic laboratory notebook]]s (ELNs) have expanded the utility of the paper [[laboratory notebook]] beyond that of a simple record keeping tool. Open ELNs offer additional benefits to the scientific community, including increased transparency, reproducibility, and [[Data integrity|integrity]]. A key element underpinning these benefits is facile and expedient knowledge sharing which aids communication and collaboration. In previous projects, we have used [[LabTrove]] and [[Vendor:LabArchives, LLC|LabArchives]] as open ELNs, in partnership with GitHub (an open-source web-based platform originally developed for collaborative coding) for communication and discussion. Here we present our personal experiences using GitHub as the central platform for many aspects of the scientific process ... ('''[[Journal:GitHub as an open electronic laboratory notebook for real-time sharing of knowledge and collaboration|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 18–24:</h2> | |||
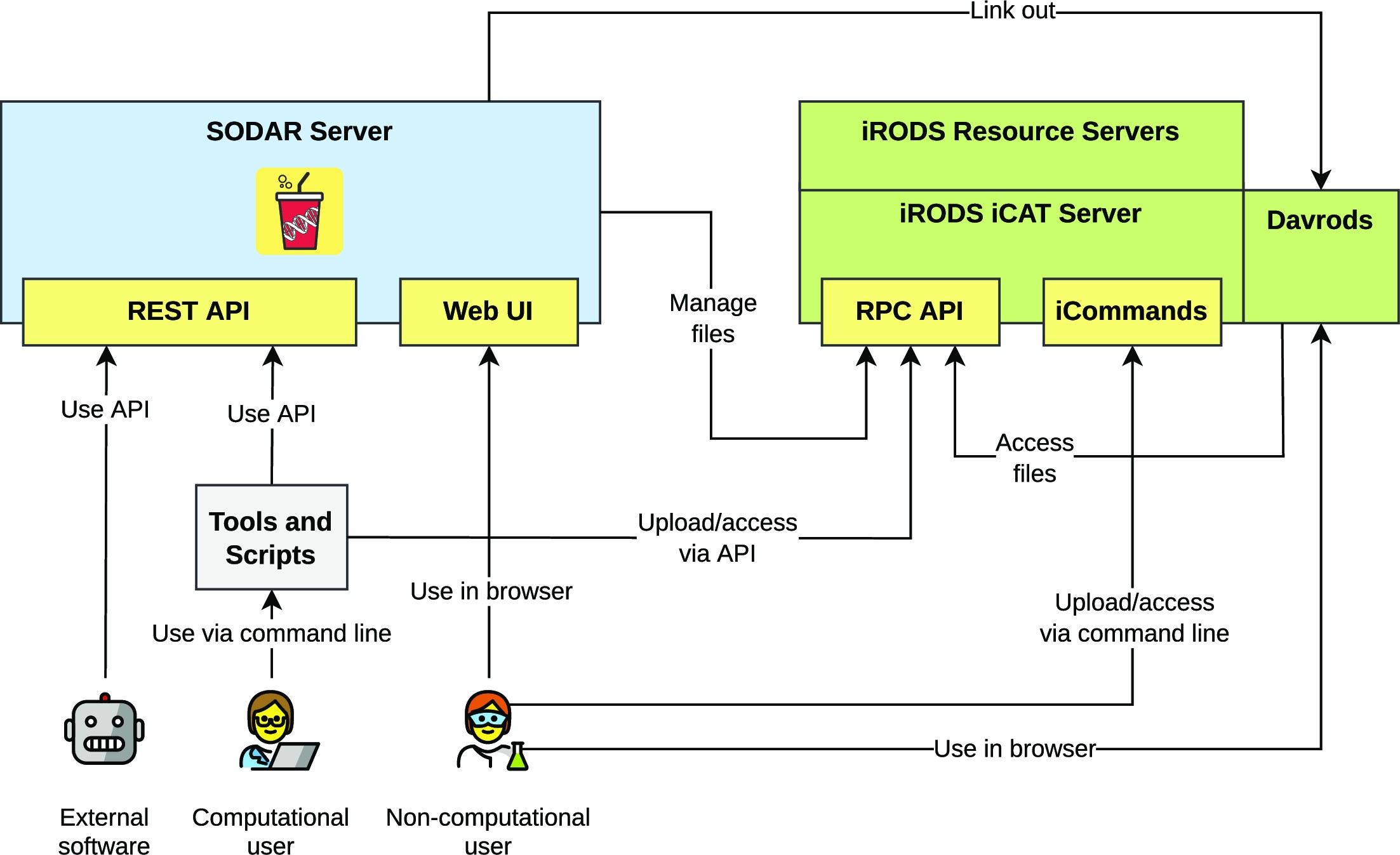
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Nieminen GigaScience2023 12.jpeg|240px]]</div> | |||
'''"[[Journal:SODAR: Managing multiomics study data and metadata|SODAR: Managing multiomics study data and metadata]]"''' | |||
Scientists employing omics in [[Life sciences industry|life science]] studies face challenges such as the modeling of multiassay studies, recording of all relevant parameters, and managing many [[Sample (material)|samples]] with their [[metadata]]. They must manage many large files that are the results of the assays or subsequent computation. Users with diverse backgrounds, ranging from computational scientists to wet-lab scientists, have dissimilar needs when it comes to data access, with programmatic interfaces being favored by the former and graphical ones by the latter. We introduce SODAR, the system for [[omics]] data access and retrieval. SODAR is a software package that addresses these challenges by providing a web-based graphical user interface (GUI) for managing multiassay studies and describing them using the ISA (Investigation, Study, Assay) data model and the ISA-Tab file format ... ('''[[Journal:SODAR: Managing multiomics study data and metadata|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 11–17:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Chang HealthInfoRes2023 29-4.png|240px]]</div> | |||
'''"[[Journal:Benefits of information technology in healthcare: Artificial intelligence, internet of things, and personal health records|Benefits of information technology in healthcare: Artificial intelligence, internet of things, and personal health records]]"''' | |||
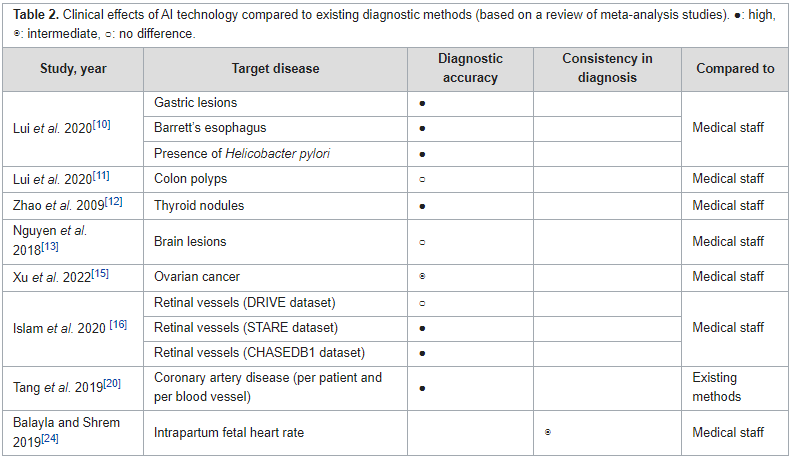
Systematic evaluations of the benefits of [[health information technology]] (HIT) play an essential role in enhancing healthcare [[Quality (business)|quality]] by improving outcomes. However, there is limited empirical evidence regarding the benefits of IT adoption in healthcare settings. This study aimed to review the benefits of [[artificial intelligence]] (AI), the [[internet of things]] (IoT), and [[personal health record]]s (PHR), based on scientific evidence. The literature published in peer-reviewed journals between 2016 and 2022 was searched for systematic reviews and meta-analysis studies using the PubMed, Cochrane, and Embase databases. Manual searches were also performed using the reference lists of systematic reviews and eligible studies from major [[health informatics]] journals. The benefits of each HIT were assessed from multiple perspectives across four outcome domains ... ('''[[Journal:Benefits of information technology in healthcare: Artificial intelligence, internet of things, and personal health records|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: March 04–10:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Villegas-Pérez Foods23 12-22.png|240px]]</div> | |||
'''"[[Journal:A quality assurance discrimination tool for the evaluation of satellite laboratory practice excellence in the context of European regulatory meat inspection for Trichinella spp.|A quality assurance discrimination tool for the evaluation of satellite laboratory practice excellence in the context of European regulatory meat inspection for ''Trichinella spp.'']]"''' | |||
[[Trichinosis|Trichinellosis]] is a parasitic foodborne zoonotic disease transmitted by ingestion of raw or undercooked meat containing the first larval stage (L1) of the nematode. To ensure the [[Quality (business)|quality]] and safety of food intended for human consumption, meat inspection for detection of ''Trichinella'' spp. larvae is a mandatory procedure per European Union (E.U.) regulations. The implementation of [[quality assurance]] (QA) practices in [[Laboratory|laboratories]] that are responsible for ''Trichinella'' spp. detection is essential given that the detection of this parasite is still a pivotal threat to public health, and it is included in List A of Annex I, Directive 2003/99/EC, which determines the agents to be monitored on a mandatory basis ... ('''[[Journal:A quality assurance discrimination tool for the evaluation of satellite laboratory practice excellence in the context of European regulatory meat inspection for Trichinella spp.|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 26–March 03:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Pineda-Pampliega EFSAJournal2023 20-S2.png|240px]]</div> | |||
'''"[[Journal:Developing a framework for open and FAIR data management practices for next generation risk- and benefit assessment of fish and seafood|Developing a framework for open and FAIR data management practices for next generation risk- and benefit assessment of fish and seafood]]"''' | |||
[[Risk assessment|Risk and risk–benefit assessments]] of food are complex exercises, in which access to and use of several disconnected individual stand-alone [[database]]s is required to obtain hazard and exposure information. Data obtained from such databases ideally should be in line with the [[Journal:The FAIR Guiding Principles for scientific data management and stewardship|FAIR principles]], i.e. the data must be findable, accessible, interoperable, and reusable. However, often cases are encountered when one or more of these principles are not followed. In this project, we set out to assess if existing commonly used databases in risk assessment are in line with the FAIR principles ... ('''[[Journal:Developing a framework for open and FAIR data management practices for next generation risk- and benefit assessment of fish and seafood|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 19–25:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Henke JMIRMedInfo2023 11.png|240px]]</div> | |||
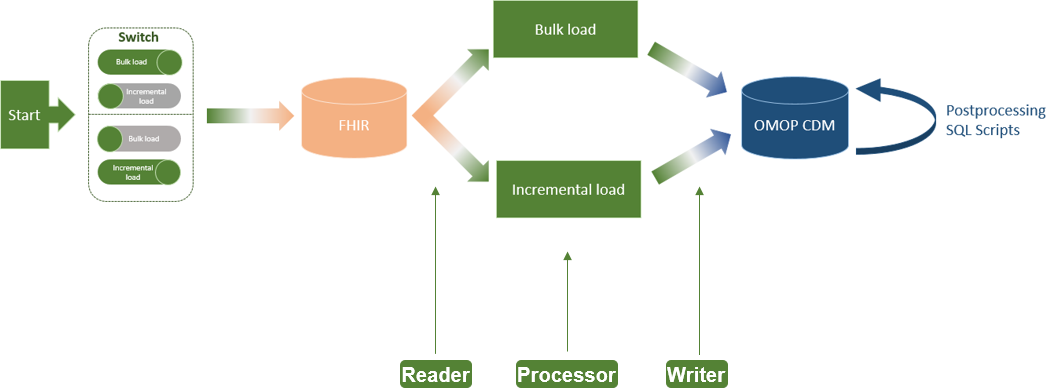
'''"[[Journal:An extract-transform-load process design for the incremental loading of German real-world data based on FHIR and OMOP CDM: Algorithm development and validation|An extract-transform-load process design for the incremental loading of German real-world data based on FHIR and OMOP CDM: Algorithm development and validation]]"''' | |||
In the Medical Informatics in Research and Care in University Medicine (MIRACUM) consortium, an IT-based clinical trial recruitment support system was developed based on the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM). Currently, OMOP CDM is populated with German Fast Healthcare Interoperability Resources (FHIR) data using an extract-transform-load (ETL) process, which was designed as a bulk load. However, the computational effort that comes with an everyday full load is not efficient for daily recruitment ... ('''[[Journal:An extract-transform-load process design for the incremental loading of German real-world data based on FHIR and OMOP CDM: Algorithm development and validation|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 12–18:</h2> | |||
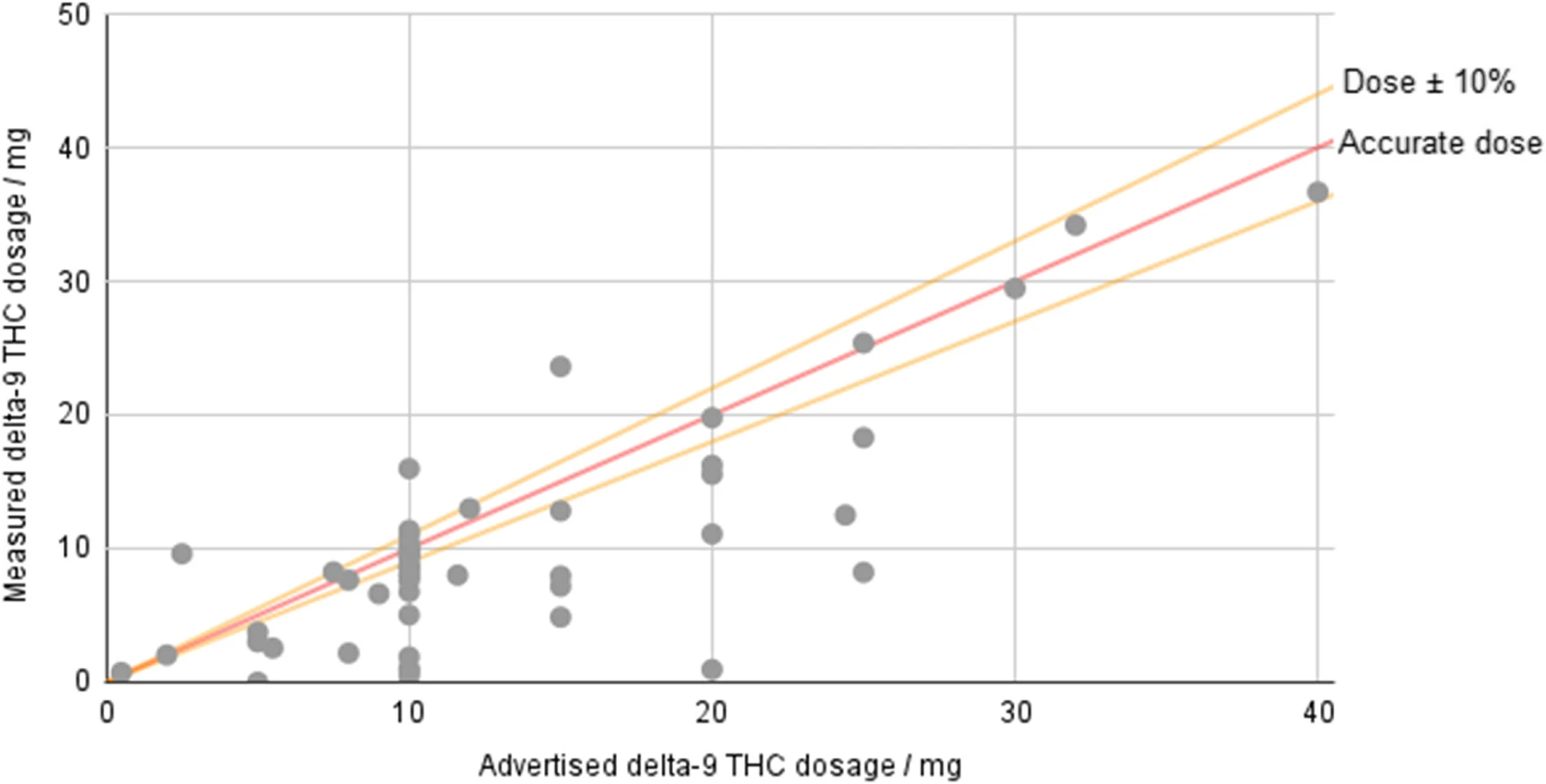
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Johnson JofCannRes23 5.png|240px]]</div> | |||
'''"[[Journal:Potency and safety analysis of hemp-derived delta-9 products: The hemp vs. cannabis demarcation problem|Potency and safety analysis of hemp-derived delta-9 products: The hemp vs. cannabis demarcation problem]]"''' | |||
[[Hemp]]-derived [[Tetrahydrocannabinol|delta-9-tetrahydrocannabinol]] (Δ<sup>9</sup>-THC) products are freely available for sale across much of the USA, but the federal legislation allowing their sale places only minimal requirements on companies. Products must contain no more than 0.3% Δ<sup>9</sup>-THC by dry weight, but no limit is placed on overall dosage, and there is no requirement that products derived from hemp-based Δ<sup>9</sup>-THC be tested. However, some states—such as Colorado—specifically prohibit products created by “chemically modifying” a natural hemp component. Fifty-three hemp-derived Δ<sup>9</sup>-THC products were ordered and submitted to InfiniteCAL [[laboratory]] for analysis ... ('''[[Journal:Potency and safety analysis of hemp-derived delta-9 products: The hemp vs. cannabis demarcation problem|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 05–11:</h2> | |||
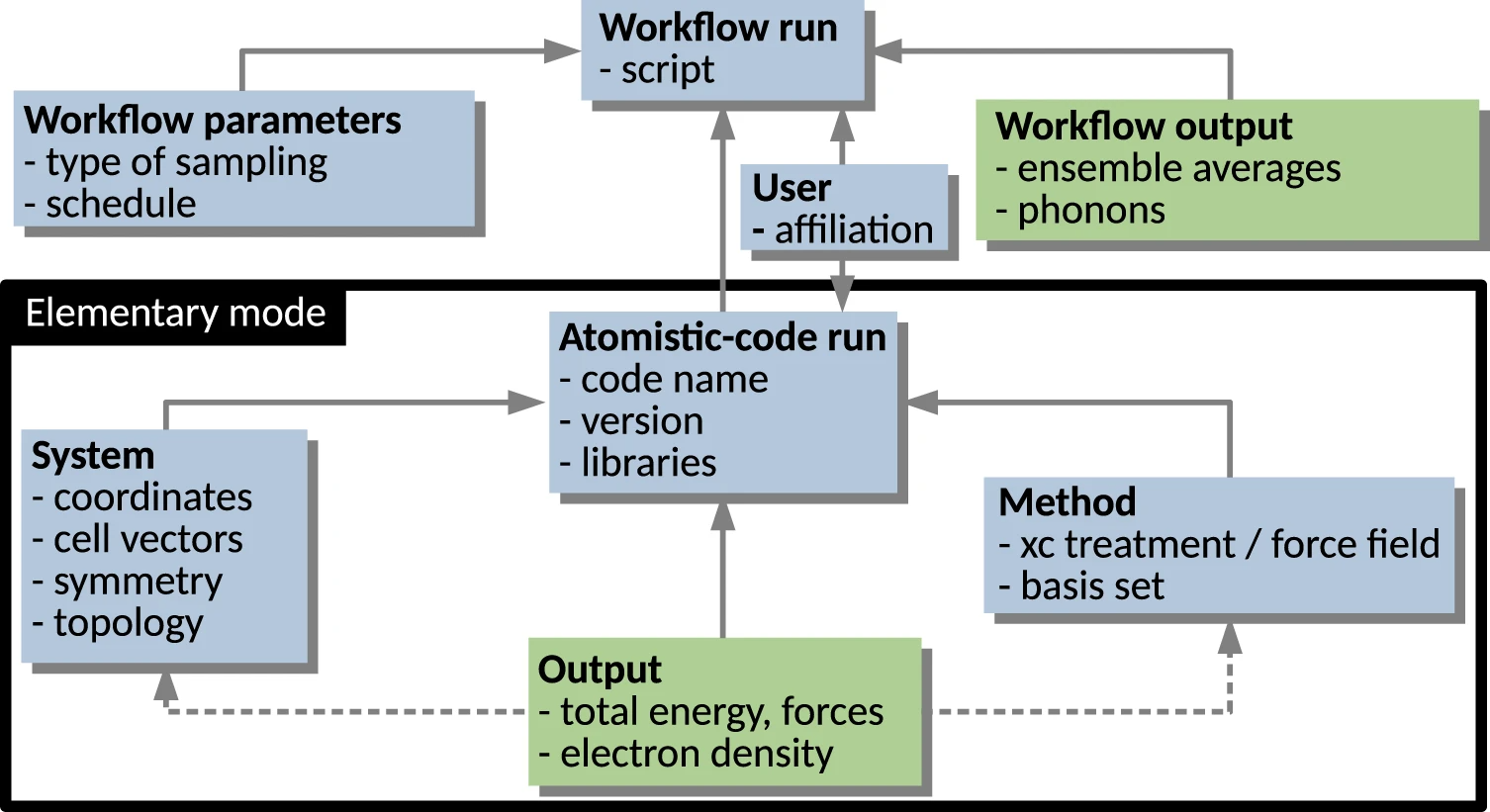
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Sbailò npjCompMat22 8.png|240px]]</div> | |||
'''"[[Journal:The NOMAD Artificial Intelligence Toolkit: Turning materials science data into knowledge and understanding|The NOMAD Artificial Intelligence Toolkit: Turning materials science data into knowledge and understanding]]"''' | |||
We present the Novel Materials Discovery (NOMAD) [[Artificial intelligence|Artificial Intelligence]] (AI) Toolkit, a web-browser-based infrastructure for the interactive AI-based analysis of [[materials science]] data under FAIR (findable, accessible, interoperable, and reusable) data principles. The AI Toolkit readily operates on FAIR data stored in the central server of the NOMAD Archive, the largest database of materials science data worldwide, as well as locally stored, user-owned data. The NOMAD Oasis, a local, stand-alone server can also be used to run the AI Toolkit. By using [[Jupyter Notebook]]s that run in a web-browser, the NOMAD data can be queried and accessed; [[data mining]], [[machine learning]] (ML), and other AI techniques can then be applied to analyze them ... ('''[[Journal:The NOMAD Artificial Intelligence Toolkit: Turning materials science data into knowledge and understanding|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 29–February 04:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Naphade JofClinDiagRes2023 17-2.jpg|240px]]</div> | |||
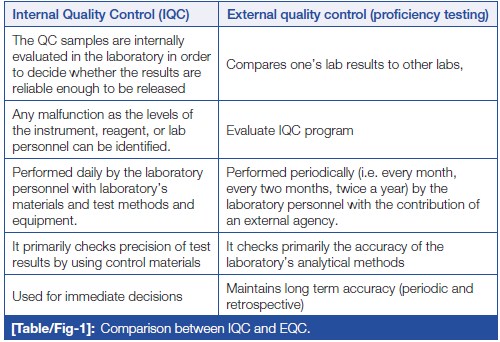
'''"[[Journal:Quality control in the clinical biochemistry laboratory: A glance|Quality control in the clinical biochemistry laboratory: A glance]]"''' | |||
[Quality control]] (QC) is a process, designed to ensure reliable test results. It is part of overall [[laboratory]] quality management in terms of accuracy, reliability, and timeliness of reported test results. Two types of QC are exercised in [[Clinical chemistry|clinical biochemistry]]: internal QC (IQC) and external [[quality assurance]] (QA). IQC represents the quality methods performed every day by laboratory personnel with the laboratory’s materials and equipment. It primarily checks the precision (i.e., repeatability or reproducibility) of the test method. External quality assurance service (EQAS) is performed periodically (i.e., every month, every two months, twice a year) by the laboratory personnel, who primarily are checking the accuracy of the laboratory’s analytical methods ... ('''[[Journal:Quality control in the clinical biochemistry laboratory: A glance|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 22–28:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Ghiringhelli SciData23 10.png|240px]]</div> | |||
'''"[[Journal:Shared metadata for data-centric materials science|Shared metadata for data-centric materials science]]"''' | |||
The expansive production of data in [[materials science]], as well as their widespread [[Data sharing|sharing]] and repurposing, requires educated support and stewardship. In order to ensure that this need helps rather than hinders scientific work, the implementation of the [[Journal:The FAIR Guiding Principles for scientific data management and stewardship|FAIR data principles]] (that ask for data and information to be findable, accessible, interoperable, and reusable) must not be too narrow. At the same time, the wider materials science community ought to agree on the strategies to tackle the challenges that are specific to its data, both from computations and experiments. In this paper, we present the result of the discussions held at the workshop on “Shared Metadata and Data Formats for Big-Data Driven Materials Science.” ... ('''[[Journal:Shared metadata for data-centric materials science|Full article...]]''')<br /> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 15–21:</h2> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Jadhav IntJofMolSci23 24-9.png|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Jadhav IntJofMolSci23 24-9.png|240px]]</div> | ||
'''"[[Journal:A metabolomics and big data approach to cannabis authenticity (authentomics)|A metabolomics and big data approach to cannabis authenticity (authentomics)]]"''' | '''"[[Journal:A metabolomics and big data approach to cannabis authenticity (authentomics)|A metabolomics and big data approach to cannabis authenticity (authentomics)]]"''' | ||
Latest revision as of 15:03, 17 June 2024
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 - 2018 - 2019 - 2020 - 2021 - 2022 - 2023 - 2024 |
Featured article of the week archive - 2024
Welcome to the LIMSwiki 2024 archive for the Featured Article of the Week.
Featured article of the week: June 10–16:"Ten simple rules for managing laboratory information" Information is the cornerstone of research, from experimental data/metadata and computational processes to complex inventories of reagents and equipment. These 10 simple rules discuss best practices for leveraging laboratory information management systems (LIMS) to transform this large information load into useful scientific findings. The development of mathematical models that can predict the properties of biological systems is the holy grail of computational biology. Such models can be used to test biological hypotheses, guide the development of biomanufactured products, engineer new systems meeting user-defined specifications, and much more ... (Full article...)
|