Difference between revisions of "Journal:Simultaneous quantification of 17 cannabinoids in cannabis inflorescence by liquid chromatography–mass spectrometry"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (→Notes: Cats) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 18: | Line 18: | ||
|website = [https://www.mdpi.com/2297-8739/9/4/85/htm https://www.mdpi.com/2297-8739/9/4/85/htm] | |website = [https://www.mdpi.com/2297-8739/9/4/85/htm https://www.mdpi.com/2297-8739/9/4/85/htm] | ||
|download = [https://www.mdpi.com/2297-8739/9/4/85/pdf?version=1648468114 https://www.mdpi.com/2297-8739/9/4/85/pdf] (PDF) | |download = [https://www.mdpi.com/2297-8739/9/4/85/pdf?version=1648468114 https://www.mdpi.com/2297-8739/9/4/85/pdf] (PDF) | ||
}} | }} | ||
==Abstract== | ==Abstract== | ||
| Line 31: | Line 25: | ||
==Introduction== | ==Introduction== | ||
''[[Cannabis sativa]]'' L. (''[[Cannabis]]'') is one of the oldest domesticated plants, cultivated for the purpose of food, fiber, [[Psychoactive drug|psychoactives]], and [[Cannabis (drug)|medicine]]. In recent years, its medicinal properties have gained a renewed interest given its therapeutic potential in the treatment of a variety of disease conditions. To date, over 100 different [[cannabinoid]]s have been identified, with [[Tetrahydrocannabinol|Δ<sup>9</sup>-tetrahydrocannabinol]] (THC), well known for its psychoactive properties, and others, such as [[cannabidiol]] (CBD), recognized for their therapeutic and medicinal value. | ''[[Cannabis sativa]]'' L. (''[[Cannabis]]'') is one of the oldest domesticated plants, cultivated for the purpose of food, fiber, [[Psychoactive drug|psychoactives]], and [[Cannabis (drug)|medicine]]. In recent years, its medicinal properties have gained a renewed interest given its therapeutic potential in the treatment of a variety of disease conditions. To date, over 100 different [[cannabinoid]]s have been identified, with [[Tetrahydrocannabinol|Δ<sup>9</sup>-tetrahydrocannabinol]] (THC), well known for its psychoactive properties, and others, such as [[cannabidiol]] (CBD), recognized for their therapeutic and medicinal value.<ref>{{Cite book |date=2017 |editor-last=Chandra |editor-first=Suman |editor2-last=ElSohly |editor2-first=Mahmoud A. |editor3-last=Lata |editor3-first=Hemant |title=Cannabis sativa L. - Botany and Biotechnology |edition=1st ed. 2017 |publisher=Springer International Publishing : Imprint: Springer |place=Cham |isbn=978-3-319-54564-6}}</ref><ref name=":0">{{Cite journal |last=Clarke |first=Robert C. |last2=Merlin |first2=Mark D. |date=2016-11-01 |title=Cannabis Domestication, Breeding History, Present-day Genetic Diversity, and Future Prospects |url=https://www.tandfonline.com/doi/full/10.1080/07352689.2016.1267498 |journal=Critical Reviews in Plant Sciences |language=en |volume=35 |issue=5-6 |pages=293–327 |doi=10.1080/07352689.2016.1267498 |issn=0735-2689}}</ref> | ||
Historically, cannabis [[Sample (material)|samples]] were analyzed mostly in a legal or regulatory context for the purpose of determining Δ<sup>9</sup>-THC content. However, as more cannabinoids were identified and their pharmacological potential was examined, the quantification of cannabinoids other than Δ<sup>9</sup>-THC gained importance. Additional cannabinoids of interest now include [[Tetrahydrocannabinolic acid|Δ<sup>9</sup>-tetrahydrocannabinolic acid]] (THCA), [[cannabidiol]] (CBD), [[cannabidiolic acid]] (CBDA), [[cannabigerol]] (CBG), [[cannabigerolic acid]] (CBGA), [[Tetrahydrocannabivarin|Δ<sup>9</sup>-tetrahydrocannabivarin]] (THCV), Δ<sup>9</sup>-tetrahydrocannabivarinic acid (THCVA), [[cannabidivarin]] (CBDV), and cannabidivarinic acid (CBDVA). | Historically, cannabis [[Sample (material)|samples]] were analyzed mostly in a legal or regulatory context for the purpose of determining Δ<sup>9</sup>-THC content. However, as more cannabinoids were identified and their pharmacological potential was examined, the quantification of cannabinoids other than Δ<sup>9</sup>-THC gained importance. Additional cannabinoids of interest now include [[Tetrahydrocannabinolic acid|Δ<sup>9</sup>-tetrahydrocannabinolic acid]] (THCA), [[cannabidiol]] (CBD), [[cannabidiolic acid]] (CBDA), [[cannabigerol]] (CBG), [[cannabigerolic acid]] (CBGA), [[Tetrahydrocannabivarin|Δ<sup>9</sup>-tetrahydrocannabivarin]] (THCV), Δ<sup>9</sup>-tetrahydrocannabivarinic acid (THCVA), [[cannabidivarin]] (CBDV), and cannabidivarinic acid (CBDVA).<ref name=":0" /><ref name=":1">{{Cite journal |last=Giese |first=Matthew W |last2=Lewis |first2=Mark A |last3=Giese |first3=Laura |last4=Smith |first4=Kevin M |date=2015-11-01 |title=Method for the Analysis of Cannabinoids and Terpenes in Cannabis |url=https://academic.oup.com/jaoac/article/98/6/1503-1522/5654599 |journal=Journal of AOAC INTERNATIONAL |language=en |volume=98 |issue=6 |pages=1503–1522 |doi=10.5740/jaoacint.15-116 |issn=1060-3271}}</ref> A recent study on virus-neutralizing capabilities of naturally occurring cannabinoids (in acid form) even found that CBDA and CBGA are capable of binding to the spike protein of SARS-CoV-2, therefore making for possible candidates for the treatment and prevention of COVID-19.<ref>{{Cite journal |last=van Breemen |first=Richard B. |last2=Muchiri |first2=Ruth N. |last3=Bates |first3=Timothy A. |last4=Weinstein |first4=Jules B. |last5=Leier |first5=Hans C. |last6=Farley |first6=Scotland |last7=Tafesse |first7=Fikadu G. |date=2022-01-28 |title=Cannabinoids Block Cellular Entry of SARS-CoV-2 and the Emerging Variants |url=https://pubs.acs.org/doi/10.1021/acs.jnatprod.1c00946 |journal=Journal of Natural Products |language=en |volume=85 |issue=1 |pages=176–184 |doi=10.1021/acs.jnatprod.1c00946 |issn=0163-3864 |pmc=PMC8768006 |pmid=35007072}}</ref> Clearly, accurate methods of quantification are needed to determine the concentrations of both major and minor cannabinoids of pharmacological potential in cannabis [[inflorescence]], the source of cannabinoids in the plant. | ||
Despite the rising demand and importance of accurate quantification of cannabinoids, the number of studies with a clear focus on analytical method development remains relatively small. This might be in part due to [[Decriminalization of non-medical cannabis in the United States|regulatory constraints]], which necessitate specific licenses to handle this scheduled drug and limit the availability of standard compounds and test samples. Due to this, the cannabis testing industry remains poorly established, with only a relatively limited number of [[Laboratory|laboratories]] offering analytical services and a lack of standardized protocols for extraction and analysis. | Despite the rising demand and importance of accurate quantification of cannabinoids, the number of studies with a clear focus on analytical method development remains relatively small. This might be in part due to [[Decriminalization of non-medical cannabis in the United States|regulatory constraints]], which necessitate specific licenses to handle this scheduled drug and limit the availability of standard compounds and test samples. Due to this, the cannabis testing industry remains poorly established, with only a relatively limited number of [[Laboratory|laboratories]] offering analytical services and a lack of standardized protocols for extraction and analysis.<ref>{{Cite journal |last=Welling |first=Matthew T. |last2=Liu |first2=Lei |last3=Hazekamp |first3=Arno |last4=Dowell |first4=Ashley |last5=King |first5=Graham J. |date=2019-02-25 |title=Developing Robust Standardised Analytical Procedures for Cannabinoid Quantification: Laying the Foundations for an Emerging Cannabis-Based Pharmaceutical Industry |url=https://www.karger.com/Article/FullText/496868 |journal=Medical Cannabis and Cannabinoids |language=en |volume=2 |issue=1 |pages=1–13 |doi=10.1159/000496868 |issn=2504-3889 |pmc=PMC8489335 |pmid=34676328}}</ref> | ||
Two review articles published recently | Two review articles published recently<ref name=":2">{{Cite journal |last=Citti |first=Cinzia |last2=Braghiroli |first2=Daniela |last3=Vandelli |first3=Maria Angela |last4=Cannazza |first4=Giuseppe |date=2018-01 |title=Pharmaceutical and biomedical analysis of cannabinoids: A critical review |url=https://linkinghub.elsevier.com/retrieve/pii/S0731708517311895 |journal=Journal of Pharmaceutical and Biomedical Analysis |language=en |volume=147 |pages=565–579 |doi=10.1016/j.jpba.2017.06.003}}</ref><ref name=":3">{{Cite journal |last=Micalizzi |first=Giuseppe |last2=Vento |first2=Federica |last3=Alibrando |first3=Filippo |last4=Donnarumma |first4=Danilo |last5=Dugo |first5=Paola |last6=Mondello |first6=Luigi |date=2021-01 |title=Cannabis Sativa L.: a comprehensive review on the analytical methodologies for cannabinoids and terpenes characterization |url=https://linkinghub.elsevier.com/retrieve/pii/S0021967320311389 |journal=Journal of Chromatography A |language=en |volume=1637 |pages=461864 |doi=10.1016/j.chroma.2020.461864}}</ref> contain critical evaluations of methods used for cannabinoid analysis and the recent trends; therefore, a detailed comparison of methods is not included here. [[Chromatography|Chromatographic]] techniques are generally used for the separation of cannabinoids. [[Chromatography#Liquid chromatography|Liquid chromatography]] (LC) has gained preference over [[gas chromatography]] (GC) as LC avoids conversion of the acid forms of cannabinoids to their neutral forms, which occurs at the high temperatures used in GC.<ref name=":4">{{Cite journal |last=Hewavitharana |first=A.K. |last2=Golding |first2=G. |last3=Tempany |first3=G. |last4=King |first4=G. |last5=Holling |first5=N. |date=2005-05-01 |title=Quantitative GC-MS Analysis of Δ9-Tetrahydrocannabinol in Fiber Hemp Varieties |url=http://academic.oup.com/jat/article/29/4/258/730079/Quantitative-GCMS-Analysis-of |journal=Journal of Analytical Toxicology |language=en |volume=29 |issue=4 |pages=258–261 |doi=10.1093/jat/29.4.258 |issn=1945-2403}}</ref> Until recently, the detection of column-separated cannabinoids was predominantly carried out using [[Spectrophotometry|UV spectrophotometry]], which provides low specificity and makes baseline separation of all cannabinoids imperative. However, complete separation of structurally similar cannabinoids, especially isomeric compounds such as Δ<sup>8</sup>-THC and Δ<sup>9</sup>-THC, is challenging.<ref name=":1" /> Due to the lack of specificity of UV detection, any unknown compound/s co-eluting at the same retention time as the target cannabinoid can cause overestimation of its concentration. | ||
To overcome this specificity problem, the detection method of choice has been changing to [[mass spectrometry]] (MS). | To overcome this specificity problem, the detection method of choice has been changing to [[mass spectrometry]] (MS).<ref name=":2" /><ref name=":3" /> As MS detection can identify molecules according to differences in mass, the chromatographic separation becomes less important. However, since some of the major cannabinoids—e.g., Δ<sup>8</sup>-THC, Δ<sup>9</sup>-THC, CBD, [[cannabichromene]] (CBC), and [[cannabicyclol]] (CBL)—have the same molecular mass, chromatographic separation is still required. Most of the methods using MS detection of cannabinoids are based on [[tandem mass spectrometry]] (MS-MS).<ref name=":2" /><ref name=":3" /> However, there is no real advantage of using this approach over basic MS, as cannabinoids of the same molar masses produce similar fragmentation patterns in the second MS event, thus not providing additional selectivity. Therefore, baseline separation by chromatography followed by simple MS detection is well suited for the routine analysis of cannabinoids. | ||
According to a recent review of cannabinoid analysis, variability in extraction (solvent/s, method, time, and temperature) significantly contributes to differential analytical results. | According to a recent review of cannabinoid analysis, variability in extraction (solvent/s, method, time, and temperature) significantly contributes to differential analytical results.<ref name=":2" /> Most of the recent analytical methods have used [[ethanol]] for extraction of both acidic and neutral cannabinoids<ref name=":1" /><ref name=":5">{{Cite web |last=ECA Foundation |date=08 April 2020 |title=German Pharmacopoeia Monograph for Cannabis Extracts |work=ECA Academy |url=https://www.gmp-compliance.org/gmp-news/german-pharmacopoeia-monograph-for-cannabis-extracts |publisher=ECA Foundation |accessdate=28 February 2022}}</ref>, although the method used for extraction varied in terms of sample to solvent ratio, the extraction technique, and the duration of extraction. Therefore, a study of the effects of these variables on the amounts of cannabinoids extracted is warranted. | ||
The aims of this study were to develop a simple yet effective method for the extraction of cannabinoids from cannabis inflorescences, and to develop a reliable, robust and simple [[liquid chromatography–mass spectrometry]] (LC–MS) method that can be used for routine analysis of 17 phytocannabinoids for which standards are available. In this study, a [[Time-of-flight mass spectrometry|time-of-flight mass spectrometer]] (TOF-MS) was used as the detector. The acquisition of spectral data was by simple MS mode rather than MS-MS because there was no advantage in using tandem MS. As many natural products laboratories are equipped with a TOF instrument (primarily used for qualitative analysis), a quantification method based on a TOF-MS would facilitate adoption of this method. The extraction conditions were optimized to develop a simple and robust method to extract cannabinoids from inflorescence, without altering the composition of cannabinoids. A separation method that uses a low flow rate enabled the use of ([[high-performance liquid chromatography]] (HPLC) rather than requiring an expensive ultra-high-performance liquid chromatography (UHPLC) system. The chromatographic separation of the cannabinoids with the same molar mass was made sufficiently robust so that small changes in column and/or mobile phase do not cause the isobaric cannabinoids to co-elute. | The aims of this study were to develop a simple yet effective method for the extraction of cannabinoids from cannabis inflorescences, and to develop a reliable, robust and simple [[liquid chromatography–mass spectrometry]] (LC–MS) method that can be used for routine analysis of 17 phytocannabinoids for which standards are available. In this study, a [[Time-of-flight mass spectrometry|time-of-flight mass spectrometer]] (TOF-MS) was used as the detector. The acquisition of spectral data was by simple MS mode rather than MS-MS because there was no advantage in using tandem MS. As many natural products laboratories are equipped with a TOF instrument (primarily used for qualitative analysis), a quantification method based on a TOF-MS would facilitate adoption of this method. The extraction conditions were optimized to develop a simple and robust method to extract cannabinoids from inflorescence, without altering the composition of cannabinoids. A separation method that uses a low flow rate enabled the use of ([[high-performance liquid chromatography]] (HPLC) rather than requiring an expensive ultra-high-performance liquid chromatography (UHPLC) system. The chromatographic separation of the cannabinoids with the same molar mass was made sufficiently robust so that small changes in column and/or mobile phase do not cause the isobaric cannabinoids to co-elute. | ||
| Line 62: | Line 56: | ||
The chromatographic separation of cannabinoids was performed using UHPLC on an Agilent 1290 Infinity II series system (Agilent Technologies, Santa Clara, CA, USA). The UHPLC was coupled to an Agilent 6545 high-resolution accurate mass (HRAM) QToF mass spectrometer equipped with a dual nebulizer jet stream electrospray source for ionization (Agilent) and controlled using Agilent MassHunter Acquisition Software, Tokyo, Japan (Version 10.1). Separation was achieved using an Agilent Poroshell C18 2.7 µm particle size, narrow bore, 15 cm column, held at 30 °C. The chromatographic analysis was performed using 0.1% (v/v) formic acid in 30% v/v methanol in water (mobile phase A), and 0.1% formic acid (v/v) in acetonitrile (mobile phase B), at a flow rate of 0.3 mL/min. The column was pre-equilibrated with 47% (v/v) A and 53% (v/v) B. After injection (of 1 µL sample or standard), the composition of mobile phase was maintained at 53% (v/v) B for the first 25 min. It was then changed from 53% (v/v) B to 70% (v/v) B over the next 15 minutes (to 40 minute mark), changed from 70% (v/v) B to 97% (v/v) B over the next two minutes (to 42 minute mark), and then held at 97% (v/v) B for one minute (to 43 minute mark). The column was then returned to the starting composition of 53% (v/v) B over the next one minute (to 44 minute mark). Finally, the column was re-equilibrated using 53% (v/v) B for six minutes (to 50 minute mark) prior to the next injection. The injection volume was kept low to avoid peak distortion (as explained in the discussion). | The chromatographic separation of cannabinoids was performed using UHPLC on an Agilent 1290 Infinity II series system (Agilent Technologies, Santa Clara, CA, USA). The UHPLC was coupled to an Agilent 6545 high-resolution accurate mass (HRAM) QToF mass spectrometer equipped with a dual nebulizer jet stream electrospray source for ionization (Agilent) and controlled using Agilent MassHunter Acquisition Software, Tokyo, Japan (Version 10.1). Separation was achieved using an Agilent Poroshell C18 2.7 µm particle size, narrow bore, 15 cm column, held at 30 °C. The chromatographic analysis was performed using 0.1% (v/v) formic acid in 30% v/v methanol in water (mobile phase A), and 0.1% formic acid (v/v) in acetonitrile (mobile phase B), at a flow rate of 0.3 mL/min. The column was pre-equilibrated with 47% (v/v) A and 53% (v/v) B. After injection (of 1 µL sample or standard), the composition of mobile phase was maintained at 53% (v/v) B for the first 25 min. It was then changed from 53% (v/v) B to 70% (v/v) B over the next 15 minutes (to 40 minute mark), changed from 70% (v/v) B to 97% (v/v) B over the next two minutes (to 42 minute mark), and then held at 97% (v/v) B for one minute (to 43 minute mark). The column was then returned to the starting composition of 53% (v/v) B over the next one minute (to 44 minute mark). Finally, the column was re-equilibrated using 53% (v/v) B for six minutes (to 50 minute mark) prior to the next injection. The injection volume was kept low to avoid peak distortion (as explained in the discussion). | ||
The mass spectrometer was used in MS1-positive mode to scan from m/z 190 to 370 for all samples at a scan rate of 1.5 spectra/s. The general parameters of the source were: capillary voltage 4000 V, nozzle voltage 0 V, fragmentor voltage 180 V, skimmer voltage 45 V and octopole RF Peak voltage 750 V, nebulizer pressure 20 psi, drying gas 10 L/minute, drying gas temperature 325 °C, sheath gas flow 12, and sheath gas temperature 400 °C. | The mass spectrometer was used in MS1-positive mode to scan from ''m/z'' 190 to 370 for all samples at a scan rate of 1.5 spectra/s. The general parameters of the source were: capillary voltage 4000 V, nozzle voltage 0 V, fragmentor voltage 180 V, skimmer voltage 45 V and octopole RF Peak voltage 750 V, nebulizer pressure 20 psi, drying gas 10 L/minute, drying gas temperature 325 °C, sheath gas flow 12, and sheath gas temperature 400 °C. | ||
===Data analysis and calculations=== | ===Data analysis and calculations=== | ||
Data analysis was performed using Agilent MassHunter Quantitative Analysis (for QToF) Software, Tokyo, Japan (Version 10.1). Each cannabinoid in a chromatogram of standard compounds was identified based on its spectrum and the retention time. A processing method was prepared to extract each peak based on the m/z value of each compound within a ±0.03 m/z window. Peaks were integrated and exported to EXCEL for calculations. External standard calibration was performed using a five-point calibration curve up to 12.5 µg/mL concentration with 1 µL injections. The regression values obtained (slope and intercept) in EXCEL were formatted to contain the same number of significant figures as the peak area values to avoid rounding errors. Standards used in calibration were used as unknowns/samples to predict concentrations and estimate the validity of the calibration. Concentrations calculated for the samples injected were checked to ensure that they were within the calibration/linear range. If not, they were re-run after dilution. After multiplying by the dilution factor, concentrations were expressed as µg/g of the sample using the exact mass of the sample used in extraction. Dilution factors (of the 5 mL extract) used for minor and major cannabinoids in this study were 2.5 and 200, respectively. A pooled sample of ground inflorescence was prepared using a mixture of high-THC and high-CBD plant material and used as the [[quality control]] (QC) sample with each batch of the samples analyzed. | Data analysis was performed using Agilent MassHunter Quantitative Analysis (for QToF) Software, Tokyo, Japan (Version 10.1). Each cannabinoid in a chromatogram of standard compounds was identified based on its spectrum and the retention time. A processing method was prepared to extract each peak based on the ''m/z'' value of each compound within a ±0.03 ''m/z'' window. Peaks were integrated and exported to EXCEL for calculations. External standard calibration was performed using a five-point calibration curve up to 12.5 µg/mL concentration with 1 µL injections. The regression values obtained (slope and intercept) in EXCEL were formatted to contain the same number of significant figures as the peak area values to avoid rounding errors. Standards used in calibration were used as unknowns/samples to predict concentrations and estimate the validity of the calibration. Concentrations calculated for the samples injected were checked to ensure that they were within the calibration/linear range. If not, they were re-run after dilution. After multiplying by the dilution factor, concentrations were expressed as µg/g of the sample using the exact mass of the sample used in extraction. Dilution factors (of the 5 mL extract) used for minor and major cannabinoids in this study were 2.5 and 200, respectively. A pooled sample of ground inflorescence was prepared using a mixture of high-THC and high-CBD plant material and used as the [[quality control]] (QC) sample with each batch of the samples analyzed. | ||
===Extraction method used for routine analysis=== | ===Extraction method used for routine analysis=== | ||
| Line 91: | Line 85: | ||
===Optimization of the extraction=== | ===Optimization of the extraction=== | ||
Ethanol has been the most common solvent of choice in recent studies of cannabinoid analysis, and the high extracting efficiency of ethanol is attributed to the high affinity of ethanol to the cannabinoid molecular structure. | Ethanol has been the most common solvent of choice in recent studies of cannabinoid analysis, and the high extracting efficiency of ethanol is attributed to the high affinity of ethanol to the cannabinoid molecular structure.<ref name=":2" /> Important advantages of using ethanol extraction for cannabinoid analysis are its general availability, low toxicity, low cost, and easy storage and handling. Furthermore, the concentrations determined by the analytical methods that used ethanol for extraction are likely in the range of concentrations used for actual extraction in formulations intended for human consumption, as ethanol is a common extraction solvent in preparations. An extraction that does not involve heat is more appropriate for cannabinoid extraction because heat can change the ratio of acid forms to neutral forms<ref name=":4" />, thereby altering the composition of cannabinoids in the sample prior to analysis. | ||
An optimization study of the extraction of cannabinoids from dried, ground cannabis inflorescence, using factorial design, found that the extraction solvent (80% methanol vs methanol-chloroform mixture), technique of extraction, and the time of extraction beyond 15 minutes did not significantly affect the amounts of cannabinoids extracted. The most significant variable that affected the amounts extracted was the sample to solvent ratio. | An optimization study of the extraction of cannabinoids from dried, ground cannabis inflorescence, using factorial design, found that the extraction solvent (80% methanol vs methanol-chloroform mixture), technique of extraction, and the time of extraction beyond 15 minutes did not significantly affect the amounts of cannabinoids extracted. The most significant variable that affected the amounts extracted was the sample to solvent ratio.<ref name=":6">{{Cite journal |last=Mudge |first=Elizabeth M. |last2=Murch |first2=Susan J. |last3=Brown |first3=Paula N. |date=2017-05 |title=Leaner and greener analysis of cannabinoids |url=http://link.springer.com/10.1007/s00216-017-0256-3 |journal=Analytical and Bioanalytical Chemistry |language=en |volume=409 |issue=12 |pages=3153–3163 |doi=10.1007/s00216-017-0256-3 |issn=1618-2642 |pmc=PMC5395585 |pmid=28233028}}</ref> In recent methods that used ethanol as the extraction solvent, the ratio (grams of dried inflorescence/mL ethanol) varied from 1/100<ref name=":5" /><ref>{{Cite journal |last=Berman |first=Paula |last2=Futoran |first2=Kate |last3=Lewitus |first3=Gil M. |last4=Mukha |first4=Dzmitry |last5=Benami |first5=Maya |last6=Shlomi |first6=Tomer |last7=Meiri |first7=David |date=2018-12 |title=A new ESI-LC/MS approach for comprehensive metabolic profiling of phytocannabinoids in Cannabis |url=http://www.nature.com/articles/s41598-018-32651-4 |journal=Scientific Reports |language=en |volume=8 |issue=1 |pages=14280 |doi=10.1038/s41598-018-32651-4 |issn=2045-2322 |pmc=PMC6155167 |pmid=30250104}}</ref> to 1/15.<ref name=":1" /> Therefore, a study of the effect of this ratio on the amounts of cannabinoids extracted was warranted in order to establish an optimum extraction procedure prior to instrumental analysis. A systematic study of the effects of solvents other than the ones studied in the previous study<ref name=":6" />, and extraction times below 15 minutes, was beyond the scope of this study. | ||
In this study, the effect of the ratio "grams of ground inflorescence / mL ethanol" on the amount of cannabinoids extracted was studied using 0.1g of dried inflorescence in 2.5, 5, and 10 mL volumes of ethanol (ratios of 1/25, 1/50, and 1/100, respectively), using triplicate extractions for each ratio. Volumes of injection of 1, 2, and 4 µL (respectively) were used to obtain the same peak area (if each treatment were to extract the same amount). Ten cannabinoids that were present in significant amounts in the sample were used for comparison (CBDV, CBDA, CBG, CBD, THCV, CBGA, THCVA, CBN, Δ9-THC, THCA). Peak area per gram of sample was used for comparison. | In this study, the effect of the ratio "grams of ground inflorescence / mL ethanol" on the amount of cannabinoids extracted was studied using 0.1g of dried inflorescence in 2.5, 5, and 10 mL volumes of ethanol (ratios of 1/25, 1/50, and 1/100, respectively), using triplicate extractions for each ratio. Volumes of injection of 1, 2, and 4 µL (respectively) were used to obtain the same peak area (if each treatment were to extract the same amount). Ten cannabinoids that were present in significant amounts in the sample were used for comparison (CBDV, CBDA, CBG, CBD, THCV, CBGA, THCVA, CBN, Δ9-THC, THCA). Peak area per gram of sample was used for comparison. | ||
| Line 104: | Line 98: | ||
The polarity of a C18 column is suitable for the separation when a low pH is used (when the acid forms are not ionized to produce highly polar carboxylates). The detector (MS) sensitivities for neutral compounds were much higher than those for acid forms at low pH. Another advantage of using low pH was, that the neutral forms of cannabinoids naturally occur at much lower concentrations compared to the acid forms. Therefore, the difference in sensitivities was well suited for analyzing a mixture of acids and neutrals without having to make additional dilutions to quantify the range of cannabinoids of interest. Formic acid (0.1%) was used to achieve a low pH of around three in the mobile phase. | The polarity of a C18 column is suitable for the separation when a low pH is used (when the acid forms are not ionized to produce highly polar carboxylates). The detector (MS) sensitivities for neutral compounds were much higher than those for acid forms at low pH. Another advantage of using low pH was, that the neutral forms of cannabinoids naturally occur at much lower concentrations compared to the acid forms. Therefore, the difference in sensitivities was well suited for analyzing a mixture of acids and neutrals without having to make additional dilutions to quantify the range of cannabinoids of interest. Formic acid (0.1%) was used to achieve a low pH of around three in the mobile phase. | ||
Organic modifiers—methanol and acetonitrile, from two different classes of selectivity—were tested in different ratios and combinations to improve the selectivity factor of the separations. | Organic modifiers—methanol and acetonitrile, from two different classes of selectivity—were tested in different ratios and combinations to improve the selectivity factor of the separations.<ref>{{Cite book |last=Skoog |first=Douglas A. |date=1985 |title=Principles of instrumental analysis |series=Saunders golden sunburst series |edition=3rd |publisher=Saunders College Pub |place=Philadelphia |isbn=978-0-03-001229-7}}</ref> The optimal separation of all 17 cannabinoids was achieved within the isocratic section at the beginning of the gradient using 53% v/v methanol/water (with 0.1% formic acid) as mobile phase A, and acetonitrile (with 0.1% formic acid) on a Poroshell C18 column with a particle size of 2.7 µm (as detailed under Section 2 above) (Figure 1). The ''m/z'' values used for detection and the retention times for each cannabinoid are listed in Table 1. Since the samples and standards were in 100% organic solvent (ethanol), and the start of the gradient was only about 53% organic (weaker eluent strength), the chromatographic peaks were prone to distortion. With the column size and flow rate used in this study, we found that peak distortion could be prevented as long as the volume of injection was kept below 4 µL. The injection volume was kept at 1 µL throughout this study. | ||
| Line 122: | Line 116: | ||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="75%" | {| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="75%" | ||
|- | |- | ||
| colspan=" | | colspan="3" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 1.''' Retention time and precursor ions used to extract and integrate chromatographic peak of each cannabinoid. A ±0.03 ''m/z'' window was used to extract spectral data. | ||
|- | |- | ||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Compound | ! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Compound | ||
| Line 202: | Line 196: | ||
|} | |} | ||
|} | |} | ||
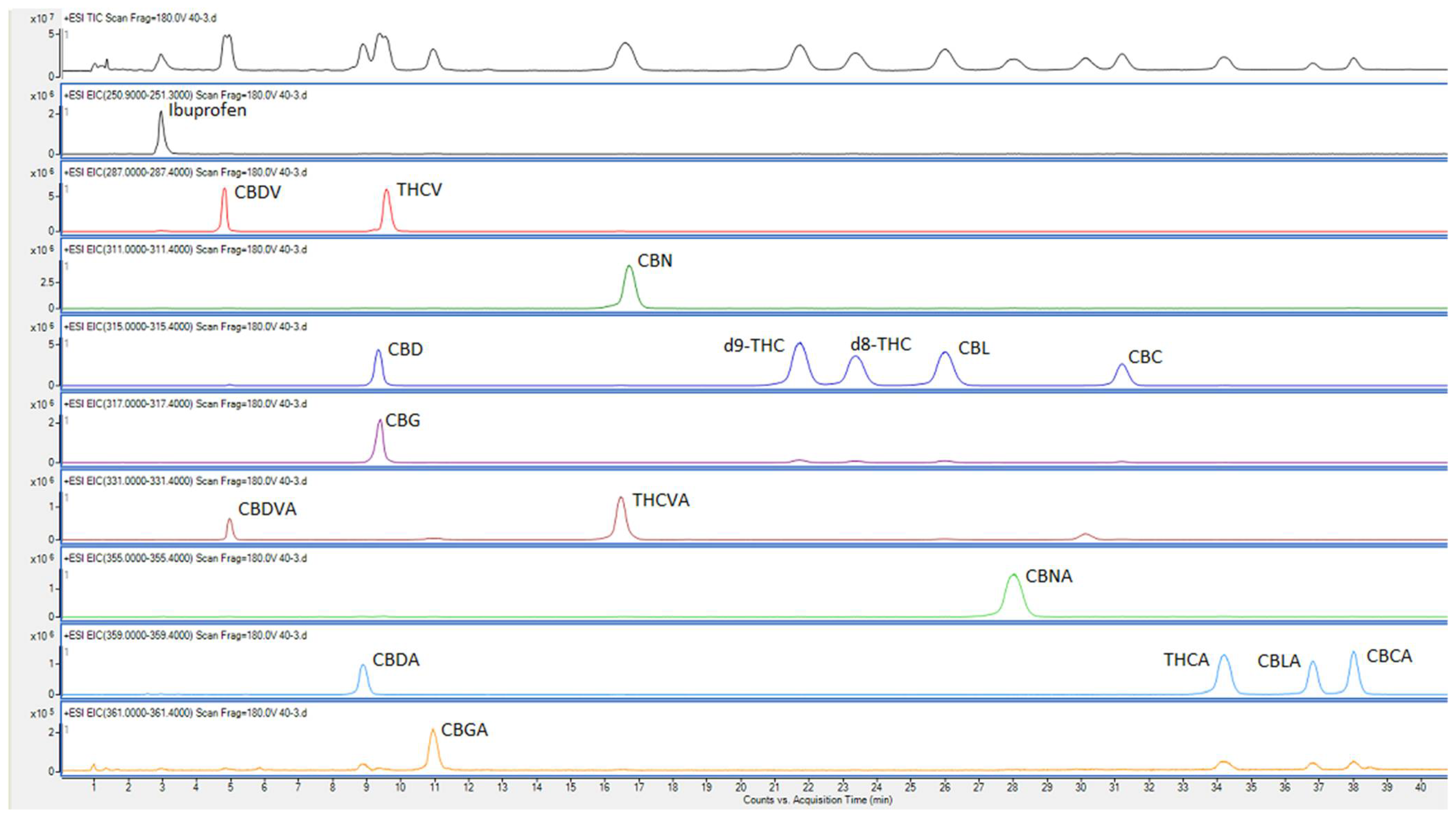
As described above, our aim was to develop an MS detection method rather than a tandem MS detection method. This was partly to be able to use an inexpensive HPLC-MS system, and partly because there is no advantage in using MS-MS detection (the isobaric cannabinoids produced the same fragments in MS-MS therefore needed to be chromatographically separated). We used the QToF and its processing software to mimic a single ion monitoring (SIM) mode of detection and quantification that is normally used in quadrupole MS systems. Data was acquired as spectra in total ion current (TIC) mode within the range of ''m/z'' 190–370, and each cannabinoid peak was extracted (as listed in Table 1) from these spectra for processing. An advantage of using the TIC mode is the possibility of detection of other compounds that have their molar masses within the range of detection used. Since we started using this method to analyze samples from other projects, we were able to detect (and tentatively identify based on literature-reported mass values) some other cannabinoids, in some samples (data not shown). This could not have been possible with a quadrupole MS instrument that is usually used for quantification. | |||
Since there was concern regarding the source temperature (325 °C) causing decarboxylation of the acid forms<ref name=":2" /><ref name=":4" /><ref>{{Cite journal |last=Wang |first=Mei |last2=Wang |first2=Yan-Hong |last3=Avula |first3=Bharathi |last4=Radwan |first4=Mohamed M. |last5=Wanas |first5=Amira S. |last6=van Antwerp |first6=John |last7=Parcher |first7=Jon F. |last8=ElSohly |first8=Mahmoud A. |last9=Khan |first9=Ikhlas A. |date=2016-12 |title=Decarboxylation Study of Acidic Cannabinoids: A Novel Approach Using Ultra-High-Performance Supercritical Fluid Chromatography/Photodiode Array-Mass Spectrometry |url=http://www.liebertpub.com/doi/10.1089/can.2016.0020 |journal=Cannabis and Cannabinoid Research |language=en |volume=1 |issue=1 |pages=262–271 |doi=10.1089/can.2016.0020 |issn=2378-8763 |pmc=PMC5549281 |pmid=28861498}}</ref>, the presence of peaks for neutral forms were checked at the retention times of the acid forms. As evident from Figure 1, they were absent, confirming that the acid forms were not decarboxylated in MS. In the case of CBDA and CBD, which have very similar retention times, this was established by running only CBDA standard, and confirming the absence of a peak at ''m/z'' of 315.2 (corresponding to CBD). | |||
===Data analysis and processing=== | |||
The extract ion chromatography (EIC) option in Agilent MassHunter Quantitative Analysis (for QToF) software (Version 10.1) was used to extract peaks. We used a window of ±0.03 ''m/z'' for EIC in this software, seeking to prevent MS line overlapping between CBD and CBG, which have the same retention time although they are 2 ''m/z'' value apart. Since the sensitivities for (abundant) acid forms were deliberately suppressed (relative to the neutral forms), a single dilution of the extract by 2.5 times allowed all minor cannabinoid levels to fall within the calibration range. The extracts were required to be diluted by 200 times to make the concentrations of the major cannabinoids (CBD, CBDA, THC, THCA) to fall within the calibration/linear range. We were able to quantify all 17 cannabinoids using only two dilutions because of the use of low pH elution to improve the sensitivities of neutral forms relative to those of acid forms, and because of the use of a low flowrate (therefore lower peak heights). | |||
Matrix effects (mostly by causing ion-suppression) are a major problem that impair the accuracy of LC-MS data.<ref name=":7">{{Cite journal |last=Taylor |first=Paul J. |date=2005-04 |title=Matrix effects: the Achilles heel of quantitative high-performance liquid chromatography–electrospray–tandem mass spectrometry |url=https://linkinghub.elsevier.com/retrieve/pii/S0009912004003364 |journal=Clinical Biochemistry |language=en |volume=38 |issue=4 |pages=328–334 |doi=10.1016/j.clinbiochem.2004.11.007}}</ref><ref name=":8">{{Cite journal |last=Hewavitharana |first=Amitha K. |date=2011-01 |title=Matrix matching in liquid chromatography–mass spectrometry with stable isotope labelled internal standards—Is it necessary? |url=https://linkinghub.elsevier.com/retrieve/pii/S0021967310016213 |journal=Journal of Chromatography A |language=en |volume=1218 |issue=2 |pages=359–361 |doi=10.1016/j.chroma.2010.11.047}}</ref><ref name=":9">{{Cite journal |last=Hewavitharana, A.K.; Shawn, P.N.; Smyth, H.D.C. et al. |date=July 2021 |title=The Importance of Complete Overlapping of Analyte and Internal Standard Peaks in Eliminating Matrix Effects with Liquid Chromatography–Mass Spectrometry (LC–MS) |url=https://www.chromatographyonline.com/view/the-importance-of-complete-overlapping-of-analyte-and-internal-standard-peaks-in-eliminating-matrix-effects-with-liquid-chromatography-mass-spectrometry-lc-ms- |journal=LCGC North America |volume=39 |issue=7 |pages=335–38}}</ref> As matrix effects cannot be completely eliminated in practice, the data is corrected for matrix effects by running a co-eluting internal standard for each analyte and subsequent calibration using internal standard method.<ref name=":7" /><ref name=":8" /> In our work, we did not correct for matrix effects because co-eluting compounds (stable isotope labelled analogues of the cannabinoids) were not yet commercially available for most of the cannabinoids. When available, they can easily be incorporated into to the current method, and the calibration can be carried out using an internal standard method. Until then, the data is not completely free of matrix effects. However, using lengthier retention times as well as achieving high resolution of all peaks in our method ensure minimal matrix effects because the likelihood of co-elution of matrix compounds with the peaks of interest is minimized. In addition, the high dilutions used, especially for the major cannabinoids (0.1 g sample diluted to 1 L), ensured negligible matrix effects on their concentration data.<ref name=":8" /> The high sensitivity of the method enables the detection of very low concentrations, and therefore high dilution to minimize matrix effects was possible. | |||
Using an internal standard that does not co-elute with the analyte is not an option in addressing LC-MS matrix effects.<ref name=":9" /> This is because the matrix effects at each time point of the chromatogram are different for different matrices. This was the reason that the internal standard (Ibuprofen) used in method development was not used in actual calculations. However, if a study were undertaken using all samples containing the same matrix (e.g., studying the effect of some variable on the same plant matrix), Ibuprofen could be used as the internal standard to eliminate procedural errors (but with no effect on matrix effects). | |||
===Method validation=== | |||
The LOD and LOQ, estimated based on 3× and 10× noise, respectively, ranged from 9 to 1734 picograms, as shown in Table 2. In general, acid forms of cannabinoids have higher LOD values (i.e., lower sensitivities) relative to the neutral forms. As discussed above, the suppression of sensitivities of acid forms (that are more abundant than the neutral forms) was deliberate in order to fit the MS responses of acid forms within the calibration range. In addition, Table 2 shows LOD and LOQ values calculated based on the concentrations of the extract that was injected to the column (with 1 µL volume of injection), and the concentrations in dried, ground inflorescence (based on 2.5-fold dilution of the 5 mL extract). | |||
{| | |||
| style="vertical-align:top;" | | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="75%" | |||
|- | |||
| colspan="8" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 2.''' Linearity, detection limits, and quantification limits for 17 cannabinoids and Ibuprofen. * On column amounts; ** Concentration in extract; *** Concentration in sample. | |||
|- | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Compound | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Linearity (r<sup>2</sup>) and (Upper limit, in µg/mL) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |LOD * (pg) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |LOD ** (µg/mL) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |LOD *** (µg/g) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |LOQ * (pg) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |LOQ ** (µg/mL) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |LOQ *** (µg/g) | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Ibuprofen | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.998 (10.0) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" | - | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" | - | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" | - | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" | - | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" | - | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" | - | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDV | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.998 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |11.0 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.011 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1.37 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |36.6 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.037 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |4.57 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCV | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |9.43 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.009 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1.18 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |31.4 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.031 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |3.93 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBN | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |72.9 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.073 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |9.11 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |243 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.243 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |30.4 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBD | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |17.4 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.017 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |2.17 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |57.9 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.058 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |7.24 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Δ<sup>8</sup>-THC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |31.4 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.031 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |3.93 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |105 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.011 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |13.1 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Δ<sup>9</sup>-THC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |23.4 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.023 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |2.93 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |78.2 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.078 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |9.77 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |60.8 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.061 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |7.60 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |203 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.020 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |25.3 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBL | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.998 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |43.9 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.044 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |5.49 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |146 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.146 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |18.3 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBG | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.996 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |27.0 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.027 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |3.37 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |89.9 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.090 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |11.2 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDVA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |181 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.181 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |22.6 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |604 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.604 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |75.4 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCVA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |98.3 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.098 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |12.3 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |328 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.328 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |40.9 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBNA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.998 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |199 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.199 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |24.8 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |662 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.662 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |82.7 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |125 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.125 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |15.7 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |418 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.418 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |52.2 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |73.7 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.074 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |9.21 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |246 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.246 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |30.7 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBLA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |239 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.239 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |29.8 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |795 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.795 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |99.4 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBCA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.999 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |220 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.220 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |27.5 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |735 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.735 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |91.8 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBGA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.997 (12.5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |520 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |0.520 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |65.0 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1734 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1.734 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |218 | |||
|- | |||
|} | |||
|} | |||
Table 2 also shows the linearity, along with the upper limits of concentrations. Good linearities (r<sup>2</sup> of >0.99) were obtained for all cannabinoids for the concentration ranges used for calibration. | |||
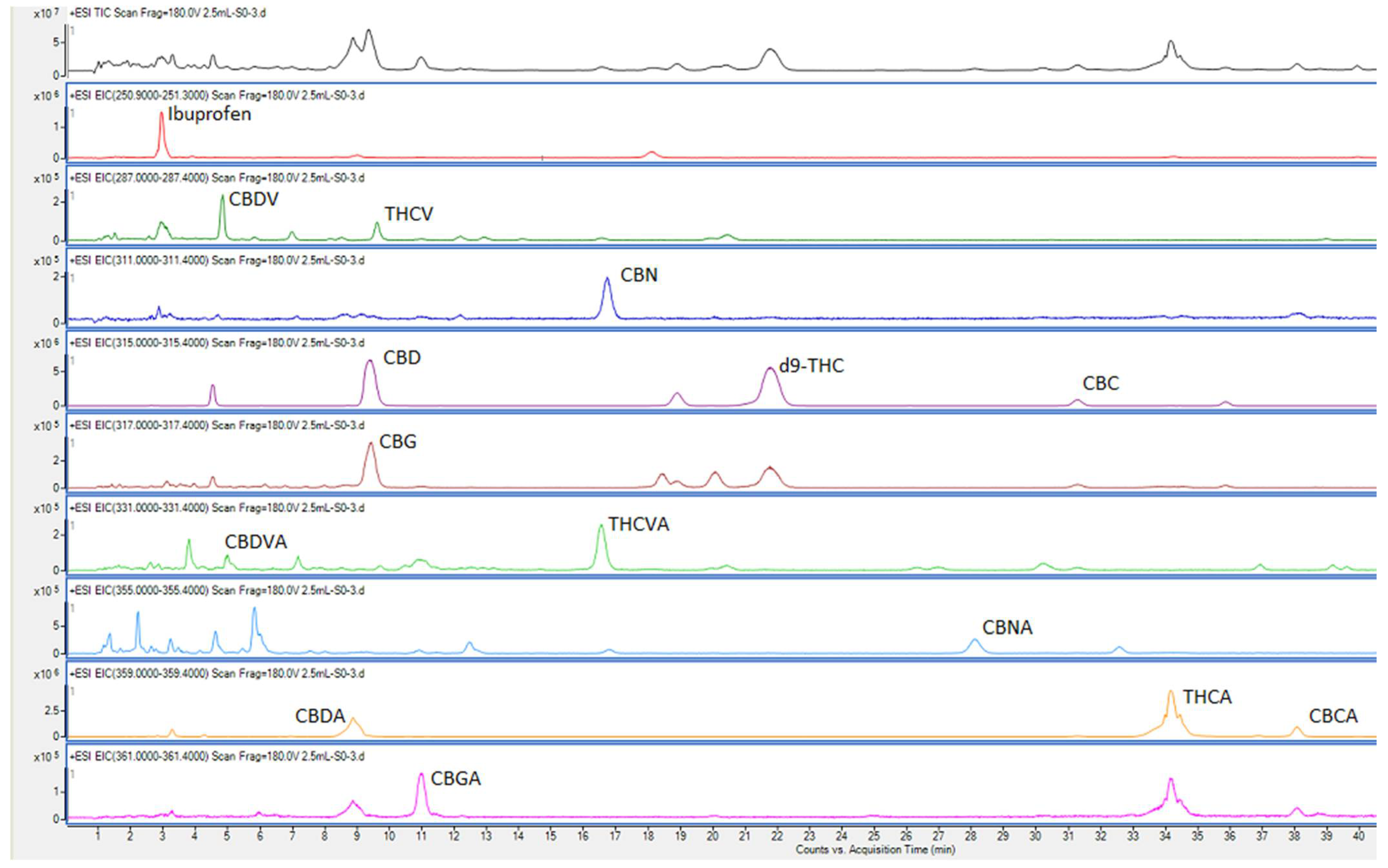
The repeatability values (''n'' = 7), determined by analyzing multiple sub-samples of the pooled ground inflorescence and expressed as percentage standard deviations, are shown in Table 3. The chromatograms for the pooled sample are shown in Figure 2. The percentage standard deviations were larger for the cannabinoids that have high LOD values/lower sensitivities. It is likely that lack of complete homogeneity of the ground samples, due to the formation of resin aggregates, contributed to the high RSD values. As suggested prior, snap-cooling the sample (in dry ice) before grinding produces homogeneous samples, although heat produced by friction during the grinding process might still cause resins to aggregate to some extent. | |||
{| | |||
| style="vertical-align:top;" | | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="75%" | |||
|- | |||
| colspan="8" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 3.''' Repeatability (as mean value with standard deviation, in column 2, and as relative standard deviation, in column 3), and recovery for each cannabinoid (''n'' = 7) in the pooled inflorescence sample. * Not detected; ** Values for ×200 diluted extract shown (values for ×2.5 diluted extracts were 381 ± 14.3, 3.76, and 29.0 ± 5.24, respectively). | |||
|- | |||
! rowspan="2" style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Compound | |||
! colspan="2" style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Repeatability | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Recovery | |||
|- | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Mean ± SD (µg/g) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |RSD (%) | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Mean ± SD (%) | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDV | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |50.5 ± 2.21 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |4.37 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |101 ± 3.15 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDVA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |144 ± 8.67 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |6.03 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |96.3 ± 6.27 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |38,850 ± 2973 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |7.65 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |94.8 ± 11.0 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBG ** | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1084 ± 91.7 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |8.46 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |97.9 ± 4.09 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBD | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |15,110 ± 973 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |6.44 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |99.0 ± 5.82 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCV | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |25.7 ± 1.46 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |5.67 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |87.3 ± 3.11 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBGA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |399 ± 20.2 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |20.2 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |108.5 ± 9.47 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCVA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |324 ± 23.0 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |7.09 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |99.4 ± 6.26 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBN | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |68.2 ± 3.66 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |5.37 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |101 ± 2.07 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Δ<sup>8</sup>-THC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND * | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" | - | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |95.0 ± 2.35 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Δ<sup>9</sup>-THC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |4748 ± 232 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |4.89 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |102 ± 4.30 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBL | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |14.7 ± 0.58 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |3.97 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |99.3 ± 3.61 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBNA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |285 ± 17.5 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |6.14 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |98.6 ± 3.32 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |770 ± 65.4 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |8.49 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |101 ± 2.87 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |43,681 ± 5080 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |11.6 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |103 ± 8.11 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBLA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |196 ± 11.0 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |5.60 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |101 ± 5.82 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBCA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |519 ± 18.8 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |3.62 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |85.2 ± 13.8 | |||
|- | |||
|} | |||
|} | |||
[[File:Fig2 Hewavitharana Separations 9-4.png|1200px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1200px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Fig. 2''' Chromatographic and mass spectrometric separation of Ibuprofen and cannabinoids in a pooled sample of cannabis inflorescence. Top chromatogram shows the total ion chromatogram and the other chromatograms are extracted at the mass of compound/s.</blockquote> | |||
|- | |||
|} | |||
|} | |||
The mean recoveries, calculated using the percent ratio of observed/spiked concentrations, are also shown in Table 3, along with their SD values (''n'' = 7). In the case of CBG, both the low-dilution (×2.5) and high-dilution (×200) produced peaks that were within the linear range. However, the recovery from low-dilution samples was about 30% compared to almost 100% recovery for the high-dilution samples. The mean concentration value was also about three times higher with the high dilution. This clearly demonstrates the ion-suppression effects on CBG, and the elimination of this effect by sample dilution, as discussed above.<ref name=":7" /> As CBG and CBD have similar retention times, the CBG signal was suppressed by CBD in high CBD samples. In the pool sample used in Table 3, the CBD concentration is >10 times that of CBG. | |||
The method was applied to quantify cannabinoids in several different samples containing high levels of CBD and high levels of THC. The concentrations of each cannabinoid quantified are listed in Table 4. | |||
{| | |||
| style="vertical-align:top;" | | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="75%" | |||
|- | |||
| colspan="3" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 4.''' Concentration ranges (µg/g) of cannabinoids found in two high-CBD and three high-THC samples. ND, not detected; <LOD, below detection limit; <LOQ, between detection and quantification limits. | |||
|- | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Compound | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |High-CBD samples | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |High-THC samples | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDV | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |61.0–82.0 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCV | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |5.95–7.26 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |38.2–107 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBN | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<LOQ | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |105–134 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBD | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |4090–28,713 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |212–382 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Δ<sup>8</sup>-THC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Δ<sup>9</sup>-THC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1705–2482 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |11,936–20,476 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBC | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |928–1095 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |153–248 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBL | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBG | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |376–470 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |519–585 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDVA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |165–216 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCVA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<LOQ | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |160–1209 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBNA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<LOD | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |342–875 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBDA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |10,258–64,992 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |245–363 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |THCA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |796–1518 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |13,479–36,420 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBLA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |196–250 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |ND | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBCA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1732–2952 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |998–1363 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CBGA | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1045–1234 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |2956–6273 | |||
|- | |||
|} | |||
|} | |||
==Conclusions== | |||
An optimised, simple extraction method followed by a reliable and simple LC-MS method, was developed and validated for the simultaneous quantification of 17 cannabinoids. The method can be used by natural products laboratories that are equipped with a TOF-MS detector. By using low flow rate, the separation was achieved at low pressure (HPLC rather than UHPLC) conditions. The disadvantage of using the low flow rate was the long run time. Detection was performed by simpler MS rather than MS-MS. Although there was no significant loss of selectivity due to using MS instead of MS-MS, the sensitivity is compromised as evident from relatively high LOD and LOQ values (µg/mL levels compared to ng/mL levels commonly achieved with MS-MS). The main advantage of using this approach is that the method can be easily adapted to use with a simple HPLC-MS (single quadrupole) system that is commonly used in routine analytical laboratories. Due to the simplicity of instrumentation, and the robustness resulting from a high resolution in the chromatography of isobaric cannabinoids, the method is well suited for routine phytocannabinoid analysis across a range of applications in a variety of laboratory settings. | |||
==Acknowledgements== | |||
We want to thank Cann Group Limited, lead industry partner on the CRC-P, for their valuable input and for providing proprietary germplasm for this research. | |||
===Author contributions=== | |||
Conceptualization A.K.H., T.K. and B.J.B.; methodology, A.K.H.; formal analysis A.K.H., F.G.-T. and M.N.; investigation, A.K.H., F.G.-T., S.P. and M.N.; writing—original draft preparation, A.K.H. and T.K.; writing—review and editing, B.J.B., F.G.-T., S.P. and M.N.; supervision, A.K.H. and T.K.; project administration, T.K. and S.P.; funding acquisition, B.J.B. and T.K. All authors have read and agreed to the published version of the manuscript. | |||
===Funding=== | |||
This project received grant funding from the Australian Government via the Department of Industry, Science, Energy and Resources, as part of the Cooperative Research Centres Project (CRC-P) program—Round 7: Growing the medicinal cannabis industry—precision farming to pharmaceuticals. | |||
===Conflicts of interest=== | |||
The authors declare no conflict of interest. | |||
==References== | ==References== | ||
| Line 210: | Line 616: | ||
<!--Place all category tags here--> | <!--Place all category tags here--> | ||
[[Category: | [[Category:LIMSwiki journal articles (added in 2022)]] | ||
[[Category: | [[Category:LIMSwiki journal articles (all)]] | ||
[[Category: | [[Category:LIMSwiki journal articles on cannabis cannabinoids]] | ||
[[Category: | [[Category:LIMSwiki journal articles on cannabis testing]] | ||
Latest revision as of 23:38, 26 December 2023
| Full article title | Simultaneous quantification of 17 cannabinoids in cannabis inflorescence by liquid chromatography–mass spectrometry |
|---|---|
| Journal | Separations |
| Author(s) | Hewavitharana, Amitha K.; Gloerfelt-Tarp, Francine; Nolan, Matthew; Barkla, Bronwyn J.; Purdy, Sarah; Kretzschmar, Tobias |
| Author affiliation(s) | Southern Cross University, New South Wales Department of Primary Industries |
| Primary contact | Email: a dot hewavitharana at pharmacy dot uq dot edu dot au |
| Year published | 2022 |
| Volume and issue | 9(4) |
| Article # | 85 |
| DOI | 10.3390/separations9040085 |
| ISSN | 2297-8739 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.mdpi.com/2297-8739/9/4/85/htm |
| Download | https://www.mdpi.com/2297-8739/9/4/85/pdf (PDF) |
Abstract
With an increasing appreciation for the unique pharmacological properties associated with distinct, individual cannabinoids of Cannabis sativa, there is demand for accurate and reliable quantification for a growing number of them. Although recent methods are based on highly selective chromatography–mass spectrometry technology, most are limited to a few cannabinoids, while relying on unnecessarily sophisticated and expensive ultra-high-performance liquid chromatography and tandem mass spectrometry. Here we report an optimized, simple extraction method followed by a reliable and simple high-performance liquid chromatography (HPLC) method for separation. The detection is performed using a time-of-flight mass spectrometer that is available in most natural products research laboratories. Due to the simplicity of instrumentation, and the robustness resulting from a high resolution in the chromatography of isobaric cannabinoids, the method is well-suited for routine phytocannabinoid analysis for a range of applications. The method was validated in terms of detection and quantification limits, repeatability, and recoveries for a total of 17 cannabinoids. Detection limits were in the range 11–520 pg when using a 1 µL sample injection volume, and the recovery percentages ranged from 85% to 108%. The validated method was subsequently applied to determine cannabinoid composition in the inflorescences of several medicinal Cannabis sativa varieties.
Keywords: cannabinoids, phytocannabinoids, LC-MS, Cannabis sativa, tetrahydrocannabinol, THC, cannabidiol, CBD
Introduction
Cannabis sativa L. (Cannabis) is one of the oldest domesticated plants, cultivated for the purpose of food, fiber, psychoactives, and medicine. In recent years, its medicinal properties have gained a renewed interest given its therapeutic potential in the treatment of a variety of disease conditions. To date, over 100 different cannabinoids have been identified, with Δ9-tetrahydrocannabinol (THC), well known for its psychoactive properties, and others, such as cannabidiol (CBD), recognized for their therapeutic and medicinal value.[1][2]
Historically, cannabis samples were analyzed mostly in a legal or regulatory context for the purpose of determining Δ9-THC content. However, as more cannabinoids were identified and their pharmacological potential was examined, the quantification of cannabinoids other than Δ9-THC gained importance. Additional cannabinoids of interest now include Δ9-tetrahydrocannabinolic acid (THCA), cannabidiol (CBD), cannabidiolic acid (CBDA), cannabigerol (CBG), cannabigerolic acid (CBGA), Δ9-tetrahydrocannabivarin (THCV), Δ9-tetrahydrocannabivarinic acid (THCVA), cannabidivarin (CBDV), and cannabidivarinic acid (CBDVA).[2][3] A recent study on virus-neutralizing capabilities of naturally occurring cannabinoids (in acid form) even found that CBDA and CBGA are capable of binding to the spike protein of SARS-CoV-2, therefore making for possible candidates for the treatment and prevention of COVID-19.[4] Clearly, accurate methods of quantification are needed to determine the concentrations of both major and minor cannabinoids of pharmacological potential in cannabis inflorescence, the source of cannabinoids in the plant.
Despite the rising demand and importance of accurate quantification of cannabinoids, the number of studies with a clear focus on analytical method development remains relatively small. This might be in part due to regulatory constraints, which necessitate specific licenses to handle this scheduled drug and limit the availability of standard compounds and test samples. Due to this, the cannabis testing industry remains poorly established, with only a relatively limited number of laboratories offering analytical services and a lack of standardized protocols for extraction and analysis.[5]
Two review articles published recently[6][7] contain critical evaluations of methods used for cannabinoid analysis and the recent trends; therefore, a detailed comparison of methods is not included here. Chromatographic techniques are generally used for the separation of cannabinoids. Liquid chromatography (LC) has gained preference over gas chromatography (GC) as LC avoids conversion of the acid forms of cannabinoids to their neutral forms, which occurs at the high temperatures used in GC.[8] Until recently, the detection of column-separated cannabinoids was predominantly carried out using UV spectrophotometry, which provides low specificity and makes baseline separation of all cannabinoids imperative. However, complete separation of structurally similar cannabinoids, especially isomeric compounds such as Δ8-THC and Δ9-THC, is challenging.[3] Due to the lack of specificity of UV detection, any unknown compound/s co-eluting at the same retention time as the target cannabinoid can cause overestimation of its concentration.
To overcome this specificity problem, the detection method of choice has been changing to mass spectrometry (MS).[6][7] As MS detection can identify molecules according to differences in mass, the chromatographic separation becomes less important. However, since some of the major cannabinoids—e.g., Δ8-THC, Δ9-THC, CBD, cannabichromene (CBC), and cannabicyclol (CBL)—have the same molecular mass, chromatographic separation is still required. Most of the methods using MS detection of cannabinoids are based on tandem mass spectrometry (MS-MS).[6][7] However, there is no real advantage of using this approach over basic MS, as cannabinoids of the same molar masses produce similar fragmentation patterns in the second MS event, thus not providing additional selectivity. Therefore, baseline separation by chromatography followed by simple MS detection is well suited for the routine analysis of cannabinoids.
According to a recent review of cannabinoid analysis, variability in extraction (solvent/s, method, time, and temperature) significantly contributes to differential analytical results.[6] Most of the recent analytical methods have used ethanol for extraction of both acidic and neutral cannabinoids[3][9], although the method used for extraction varied in terms of sample to solvent ratio, the extraction technique, and the duration of extraction. Therefore, a study of the effects of these variables on the amounts of cannabinoids extracted is warranted.
The aims of this study were to develop a simple yet effective method for the extraction of cannabinoids from cannabis inflorescences, and to develop a reliable, robust and simple liquid chromatography–mass spectrometry (LC–MS) method that can be used for routine analysis of 17 phytocannabinoids for which standards are available. In this study, a time-of-flight mass spectrometer (TOF-MS) was used as the detector. The acquisition of spectral data was by simple MS mode rather than MS-MS because there was no advantage in using tandem MS. As many natural products laboratories are equipped with a TOF instrument (primarily used for qualitative analysis), a quantification method based on a TOF-MS would facilitate adoption of this method. The extraction conditions were optimized to develop a simple and robust method to extract cannabinoids from inflorescence, without altering the composition of cannabinoids. A separation method that uses a low flow rate enabled the use of (high-performance liquid chromatography (HPLC) rather than requiring an expensive ultra-high-performance liquid chromatography (UHPLC) system. The chromatographic separation of the cannabinoids with the same molar mass was made sufficiently robust so that small changes in column and/or mobile phase do not cause the isobaric cannabinoids to co-elute.
Materials and methods
Materials
Proprietary medicinal cannabis samples of Cann Group Limited were supplied by New South Wales Department of Primary Industries (NSW DPI, Orange, Australia) under an Office of Drug Control (ODC) permit and received by Southern Cross University (SCU) under NSW Health authority. Samples of contrasting lines for THC/A and CBD/A content were used for method establishment and validation.
All cannabinoid reference standards were purchased from Novachem Pty. Ltd. (Heidelberg, Australia) as ampoules of 1,000 µg/mL solutions. The standards for (±)-Δ9-tetrahydrocannabinolic acid A (Δ9-THCA-A), tetrahydrocannabivarinic acid (THCVA), cannabidiolic acid (CBDA), cannabidivarinic acid (CBDVA), cannabigerolic acid (CBGA), cannabinolic Acid (CBNA), cannabichromenic acid (CBCA), (±)-cannabicyclol (CBL), and cannabicyclolic acid (CBLA) were supplied in acetonitrile. The standards for (−)-Δ9-tetrahydrocannabinol (Δ9-THC), (−)-Δ8-tetrahydrocannabinol (Δ8-THC), tetrahydrocannabivarin (THCV), cannabidiol (CBD), cannabidivarin (CBDV), cannabigerol (CBG), cannabinol (CBN), and cannabichromene (CBC) were supplied in methanol. Ibuprofen was purchased from Sigma-Aldrich Pty Ltd. (North Ryde, NSW, Australia). HPLC-grade ethanol (Scharlau, Scharlab S.L., Barcelona, Spain) was used for sample extractions, and LC-MS grade solvents (Honeywell Research Chemicals, Seelze, Germany) and MilliQ water (Merck Millipore, Billerica, MA, USA) were used to prepare mobile phase.
Sample preparation
Apical and mature female inflorescences were cut from the main stems, and any large stem pieces were removed. Samples were dried for approximately seven days at 24 °C under low humidity (50–58%) until a dry weight of 21% of the starting wet weight was achieved, after which they were weighed, double vacuum packed, and stored at ambient room temperature in the dark.
Two grams of inflorescence were ground to a fine powder using a 50 mL stainless steel screw top Retsch grinding canister, containing a single 20 mm stainless steel grinding ball, in a Retsch Mixer Mill MM301 (Retsch GmbH, Mettmann, Germany) at 30 rotations per second for 10 seconds. Ground samples were stored in polypropylene tubes at −20 °C until extracted for analysis (note: flash-cooling the grinding canister and ball in liquid nitrogen before adding the sample prior to grinding helped to prevent clumping of material, especially in high resin samples, thus producing a homogenous fine powder).
Optimization of the extraction
For extraction, 0.1g of ground sample was accurately weighed into a 20 mL polypropylene centrifuge tube. An aliquot of 2.50, 5.00 or 10.00 mL of 100% ethanol was accurately pipetted into the tube. Three replicates were extracted for each volume added. The mixture was sonicated in an ultrasound bath (SONICLEAN, Soniclean®, Dudley Park, Australia) at 50/60 Hz and 150 W for 15 minutes, then centrifuged at 3000 rpm (Sigma 3–16 L, Sigma Laborzentrifugen GmbH, Osterode am Harz, Germany) for 10 minutes. The supernatant was carefully removed and stored in an Eppendorf tube at −20 °C. The extract was diluted as necessary with 100% ethanol for instrumental analysis using HPLC-MS. Volumes of injection of 1, 2 and 4 µL (for 2.50, 5.00 and 10.00 mL extractions, respectively) were used to obtain the same peak area (if each treatment were to extract the same amount). Ten cannabinoids were used for comparison (CBDV, CBDA, CBG, CBD, THCV, CBGA, THCVA, CBN, Δ9-THC, THCA). The peak areas of each cannabinoid were divided by the mass of the sample, and the peak area per gram of sample was used for comparison.
Separation and detection
The chromatographic separation of cannabinoids was performed using UHPLC on an Agilent 1290 Infinity II series system (Agilent Technologies, Santa Clara, CA, USA). The UHPLC was coupled to an Agilent 6545 high-resolution accurate mass (HRAM) QToF mass spectrometer equipped with a dual nebulizer jet stream electrospray source for ionization (Agilent) and controlled using Agilent MassHunter Acquisition Software, Tokyo, Japan (Version 10.1). Separation was achieved using an Agilent Poroshell C18 2.7 µm particle size, narrow bore, 15 cm column, held at 30 °C. The chromatographic analysis was performed using 0.1% (v/v) formic acid in 30% v/v methanol in water (mobile phase A), and 0.1% formic acid (v/v) in acetonitrile (mobile phase B), at a flow rate of 0.3 mL/min. The column was pre-equilibrated with 47% (v/v) A and 53% (v/v) B. After injection (of 1 µL sample or standard), the composition of mobile phase was maintained at 53% (v/v) B for the first 25 min. It was then changed from 53% (v/v) B to 70% (v/v) B over the next 15 minutes (to 40 minute mark), changed from 70% (v/v) B to 97% (v/v) B over the next two minutes (to 42 minute mark), and then held at 97% (v/v) B for one minute (to 43 minute mark). The column was then returned to the starting composition of 53% (v/v) B over the next one minute (to 44 minute mark). Finally, the column was re-equilibrated using 53% (v/v) B for six minutes (to 50 minute mark) prior to the next injection. The injection volume was kept low to avoid peak distortion (as explained in the discussion).
The mass spectrometer was used in MS1-positive mode to scan from m/z 190 to 370 for all samples at a scan rate of 1.5 spectra/s. The general parameters of the source were: capillary voltage 4000 V, nozzle voltage 0 V, fragmentor voltage 180 V, skimmer voltage 45 V and octopole RF Peak voltage 750 V, nebulizer pressure 20 psi, drying gas 10 L/minute, drying gas temperature 325 °C, sheath gas flow 12, and sheath gas temperature 400 °C.
Data analysis and calculations
Data analysis was performed using Agilent MassHunter Quantitative Analysis (for QToF) Software, Tokyo, Japan (Version 10.1). Each cannabinoid in a chromatogram of standard compounds was identified based on its spectrum and the retention time. A processing method was prepared to extract each peak based on the m/z value of each compound within a ±0.03 m/z window. Peaks were integrated and exported to EXCEL for calculations. External standard calibration was performed using a five-point calibration curve up to 12.5 µg/mL concentration with 1 µL injections. The regression values obtained (slope and intercept) in EXCEL were formatted to contain the same number of significant figures as the peak area values to avoid rounding errors. Standards used in calibration were used as unknowns/samples to predict concentrations and estimate the validity of the calibration. Concentrations calculated for the samples injected were checked to ensure that they were within the calibration/linear range. If not, they were re-run after dilution. After multiplying by the dilution factor, concentrations were expressed as µg/g of the sample using the exact mass of the sample used in extraction. Dilution factors (of the 5 mL extract) used for minor and major cannabinoids in this study were 2.5 and 200, respectively. A pooled sample of ground inflorescence was prepared using a mixture of high-THC and high-CBD plant material and used as the quality control (QC) sample with each batch of the samples analyzed.
Extraction method used for routine analysis
A sample of 0.1g ground cannabis inflorescence was accurately weighed into a 20 mL polypropylene centrifuge tube. A 5.00 mL aliquot of 100% ethanol was accurately pipetted into the tube. The mixture was sonicated in an ultrasound bath at 50/60 Hz and 150 W for 30 minutes, then centrifuged at 3,000 rpm (Sigma 3–16 L, Germany) for 10 minutes. The supernatant was carefully removed and stored in a 2 mL Eppendorf tube at −20 °C. The extract was diluted as necessary (below) with 100% ethanol prior to the instrumental analysis using HPLC-MS.
Preparation of samples for major cannabinoids: THC, THCA, CBD, CBDA
The extract was diluted 80 times by mixing 20 µL sample extract and 1580 µL ethanol. The extract was then further diluted by mixing 400 µL of the diluted extract, 40 µL of 0.025 mg/mL Ibuprofen, and 560 µL ethanol in an HPLC vial to be used for LC-MS analysis. This resulted in a 200 times diluted extract containing 1 µg/mL Ibuprofen (internal standard).
Preparation of samples for minor cannabinoids
The extract was diluted by mixing 400 µL of the extract, 40 µL of 0.025 mg/mL Ibuprofen, and 560 µL ethanol in an HPLC vial to be used for LC-MS analysis. This resulted in a 2.5 times diluted extract containing 1 µg/mL Ibuprofen (internal standard).
Preparation of standard solutions
A 0.05 mg/mL solution of 17 cannabinoid standards was prepared by mixing 100 µL each of the 17 cannabinoid stock standards (1 mg/mL), and 300 µL of HPLC grade ethanol in a 2 mL Eppendorf tube. A 0.025 mg/mL mix of standards was prepared by diluting the 0.05 mg/mL solution and used as the working standard stock solution.
Ibuprofen was weighed into a 1.5 mL Eppendorf tube and dissolved in an appropriate volume of HPLC grade ethanol to make a 1 mg/mL stock solution. This solution was diluted with an appropriate volume of ethanol to make the 0.025 mg/mL working stock standard.
Five calibration standards were prepared by diluting the appropriate volumes of 0.025 mg/mL mix of 17 standards, and 0.025 mg/mL Ibuprofen to contain 2.5, 5, 7.5, 10, and 12.5 µg/mL cannabinoids and 1 µg/mL Ibuprofen (internal standard).
Method validation
Linearity for each cannabinoid was estimated by injecting 1µL each of five mixed standard solutions containing 0.5 to 12.5 µg/mL of the 17 cannabinoids. The linearity was assessed by plotting the peak area of each cannabinoid versus the concentration. Limit of detection (LOD) and limit of quantification (LOQ) for each cannabinoid was determined based on the peak heights (and then the concentrations) corresponding to 3× noise and 10× noise, respectively. Repeatability was estimated by analyzing seven individual 0.1 g ground cannabis inflorescence samples (that were prepared by pooling samples containing high CBD and high THC levels), followed by the calculation of relative standard deviation of the concentrations determined for each cannabinoid. Recovery was determined by using the concentrations of each cannabinoid in spiked and un-spiked extracts using seven of the individual extracts prepared for the repeatability experiment. Each extract was spiked to give an added concentration that was below the mid-point of the calibration (linear) range.
Results and discussion
Sample preparation
A significantly larger (than the amount used for analysis) amount of inflorescence (2 g) was ground in order to minimize effects of within sample variation due to the architecture/morphology of the inflorescence (containing bracts, stigma, sugar leaves, stem remnants, etc.) of varying trichome density. After homogenization by grinding, a representative 0.1 g sample was extracted for analysis.
Optimization of the extraction
Ethanol has been the most common solvent of choice in recent studies of cannabinoid analysis, and the high extracting efficiency of ethanol is attributed to the high affinity of ethanol to the cannabinoid molecular structure.[6] Important advantages of using ethanol extraction for cannabinoid analysis are its general availability, low toxicity, low cost, and easy storage and handling. Furthermore, the concentrations determined by the analytical methods that used ethanol for extraction are likely in the range of concentrations used for actual extraction in formulations intended for human consumption, as ethanol is a common extraction solvent in preparations. An extraction that does not involve heat is more appropriate for cannabinoid extraction because heat can change the ratio of acid forms to neutral forms[8], thereby altering the composition of cannabinoids in the sample prior to analysis.
An optimization study of the extraction of cannabinoids from dried, ground cannabis inflorescence, using factorial design, found that the extraction solvent (80% methanol vs methanol-chloroform mixture), technique of extraction, and the time of extraction beyond 15 minutes did not significantly affect the amounts of cannabinoids extracted. The most significant variable that affected the amounts extracted was the sample to solvent ratio.[10] In recent methods that used ethanol as the extraction solvent, the ratio (grams of dried inflorescence/mL ethanol) varied from 1/100[9][11] to 1/15.[3] Therefore, a study of the effect of this ratio on the amounts of cannabinoids extracted was warranted in order to establish an optimum extraction procedure prior to instrumental analysis. A systematic study of the effects of solvents other than the ones studied in the previous study[10], and extraction times below 15 minutes, was beyond the scope of this study.
In this study, the effect of the ratio "grams of ground inflorescence / mL ethanol" on the amount of cannabinoids extracted was studied using 0.1g of dried inflorescence in 2.5, 5, and 10 mL volumes of ethanol (ratios of 1/25, 1/50, and 1/100, respectively), using triplicate extractions for each ratio. Volumes of injection of 1, 2, and 4 µL (respectively) were used to obtain the same peak area (if each treatment were to extract the same amount). Ten cannabinoids that were present in significant amounts in the sample were used for comparison (CBDV, CBDA, CBG, CBD, THCV, CBGA, THCVA, CBN, Δ9-THC, THCA). Peak area per gram of sample was used for comparison.
According to the results of one-way ANOVA (plus a Duncan’s multiple range test for the means), sample/solvent ratio of 1/100 extracted significantly lower (p < 0.05) amounts of all cannabinoids in comparison to the other two ratios. The amounts extracted with the ratios 1/25 and 1/50 were not significantly different (at p = 0.05) for all cannabinoids. However, the precision (based on the coefficient of variance) was better with the ratio 1/50. Therefore, that ratio (0.1 g of ground inflorescence in 5 mL ethanol) was used for all extractions in this study.
Optimization of chromatographic separation and mass spectrometry
Chromatographic separation for all cannabinoids was optimized, with the main emphasis on the baseline separation of the critical isomeric pair of Δ8-THC and Δ9-THC. Chromatographic separation with good resolution was also targeted for individual cannabinoids in two groups of isobaric compounds: CBD, Δ8-THC, Δ9-THC, CBC, CBL (m/z = 315.2) and CBDA, THCA, CBLA, CBCA (m/z = 359.2). In order to conduct these separations, a column with small particle size and/or maximum possible length was important. However, because of the aim of making this separation suitable for HPLC rather than UHPLC, it was decided to use a poroshell-type column (a thin layer of porous stationary phase is coated on solid silica cores) that can produce resolutions similar to sub-2 µm body-porous stationary phase particles used in UHPLC columns, without having to use very high pressure. As the longest available LC-MS column (15 cm) was used in this separation, the high pressure required was compensated by using a low flow rate (0.3 mL/min). Although the run time could have been shortened by using a higher flow rate, the flow rate was kept low in order to operate the LC in HPLC mode rather than UHPLC mode, particularly because our aim was to produce a method that can be run on a simple HPLC system.
The polarity of a C18 column is suitable for the separation when a low pH is used (when the acid forms are not ionized to produce highly polar carboxylates). The detector (MS) sensitivities for neutral compounds were much higher than those for acid forms at low pH. Another advantage of using low pH was, that the neutral forms of cannabinoids naturally occur at much lower concentrations compared to the acid forms. Therefore, the difference in sensitivities was well suited for analyzing a mixture of acids and neutrals without having to make additional dilutions to quantify the range of cannabinoids of interest. Formic acid (0.1%) was used to achieve a low pH of around three in the mobile phase.
Organic modifiers—methanol and acetonitrile, from two different classes of selectivity—were tested in different ratios and combinations to improve the selectivity factor of the separations.[12] The optimal separation of all 17 cannabinoids was achieved within the isocratic section at the beginning of the gradient using 53% v/v methanol/water (with 0.1% formic acid) as mobile phase A, and acetonitrile (with 0.1% formic acid) on a Poroshell C18 column with a particle size of 2.7 µm (as detailed under Section 2 above) (Figure 1). The m/z values used for detection and the retention times for each cannabinoid are listed in Table 1. Since the samples and standards were in 100% organic solvent (ethanol), and the start of the gradient was only about 53% organic (weaker eluent strength), the chromatographic peaks were prone to distortion. With the column size and flow rate used in this study, we found that peak distortion could be prevented as long as the volume of injection was kept below 4 µL. The injection volume was kept at 1 µL throughout this study.
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
As described above, our aim was to develop an MS detection method rather than a tandem MS detection method. This was partly to be able to use an inexpensive HPLC-MS system, and partly because there is no advantage in using MS-MS detection (the isobaric cannabinoids produced the same fragments in MS-MS therefore needed to be chromatographically separated). We used the QToF and its processing software to mimic a single ion monitoring (SIM) mode of detection and quantification that is normally used in quadrupole MS systems. Data was acquired as spectra in total ion current (TIC) mode within the range of m/z 190–370, and each cannabinoid peak was extracted (as listed in Table 1) from these spectra for processing. An advantage of using the TIC mode is the possibility of detection of other compounds that have their molar masses within the range of detection used. Since we started using this method to analyze samples from other projects, we were able to detect (and tentatively identify based on literature-reported mass values) some other cannabinoids, in some samples (data not shown). This could not have been possible with a quadrupole MS instrument that is usually used for quantification.
Since there was concern regarding the source temperature (325 °C) causing decarboxylation of the acid forms[6][8][13], the presence of peaks for neutral forms were checked at the retention times of the acid forms. As evident from Figure 1, they were absent, confirming that the acid forms were not decarboxylated in MS. In the case of CBDA and CBD, which have very similar retention times, this was established by running only CBDA standard, and confirming the absence of a peak at m/z of 315.2 (corresponding to CBD).
Data analysis and processing
The extract ion chromatography (EIC) option in Agilent MassHunter Quantitative Analysis (for QToF) software (Version 10.1) was used to extract peaks. We used a window of ±0.03 m/z for EIC in this software, seeking to prevent MS line overlapping between CBD and CBG, which have the same retention time although they are 2 m/z value apart. Since the sensitivities for (abundant) acid forms were deliberately suppressed (relative to the neutral forms), a single dilution of the extract by 2.5 times allowed all minor cannabinoid levels to fall within the calibration range. The extracts were required to be diluted by 200 times to make the concentrations of the major cannabinoids (CBD, CBDA, THC, THCA) to fall within the calibration/linear range. We were able to quantify all 17 cannabinoids using only two dilutions because of the use of low pH elution to improve the sensitivities of neutral forms relative to those of acid forms, and because of the use of a low flowrate (therefore lower peak heights).
Matrix effects (mostly by causing ion-suppression) are a major problem that impair the accuracy of LC-MS data.[14][15][16] As matrix effects cannot be completely eliminated in practice, the data is corrected for matrix effects by running a co-eluting internal standard for each analyte and subsequent calibration using internal standard method.[14][15] In our work, we did not correct for matrix effects because co-eluting compounds (stable isotope labelled analogues of the cannabinoids) were not yet commercially available for most of the cannabinoids. When available, they can easily be incorporated into to the current method, and the calibration can be carried out using an internal standard method. Until then, the data is not completely free of matrix effects. However, using lengthier retention times as well as achieving high resolution of all peaks in our method ensure minimal matrix effects because the likelihood of co-elution of matrix compounds with the peaks of interest is minimized. In addition, the high dilutions used, especially for the major cannabinoids (0.1 g sample diluted to 1 L), ensured negligible matrix effects on their concentration data.[15] The high sensitivity of the method enables the detection of very low concentrations, and therefore high dilution to minimize matrix effects was possible.
Using an internal standard that does not co-elute with the analyte is not an option in addressing LC-MS matrix effects.[16] This is because the matrix effects at each time point of the chromatogram are different for different matrices. This was the reason that the internal standard (Ibuprofen) used in method development was not used in actual calculations. However, if a study were undertaken using all samples containing the same matrix (e.g., studying the effect of some variable on the same plant matrix), Ibuprofen could be used as the internal standard to eliminate procedural errors (but with no effect on matrix effects).
Method validation
The LOD and LOQ, estimated based on 3× and 10× noise, respectively, ranged from 9 to 1734 picograms, as shown in Table 2. In general, acid forms of cannabinoids have higher LOD values (i.e., lower sensitivities) relative to the neutral forms. As discussed above, the suppression of sensitivities of acid forms (that are more abundant than the neutral forms) was deliberate in order to fit the MS responses of acid forms within the calibration range. In addition, Table 2 shows LOD and LOQ values calculated based on the concentrations of the extract that was injected to the column (with 1 µL volume of injection), and the concentrations in dried, ground inflorescence (based on 2.5-fold dilution of the 5 mL extract).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 2 also shows the linearity, along with the upper limits of concentrations. Good linearities (r2 of >0.99) were obtained for all cannabinoids for the concentration ranges used for calibration.
The repeatability values (n = 7), determined by analyzing multiple sub-samples of the pooled ground inflorescence and expressed as percentage standard deviations, are shown in Table 3. The chromatograms for the pooled sample are shown in Figure 2. The percentage standard deviations were larger for the cannabinoids that have high LOD values/lower sensitivities. It is likely that lack of complete homogeneity of the ground samples, due to the formation of resin aggregates, contributed to the high RSD values. As suggested prior, snap-cooling the sample (in dry ice) before grinding produces homogeneous samples, although heat produced by friction during the grinding process might still cause resins to aggregate to some extent.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
The mean recoveries, calculated using the percent ratio of observed/spiked concentrations, are also shown in Table 3, along with their SD values (n = 7). In the case of CBG, both the low-dilution (×2.5) and high-dilution (×200) produced peaks that were within the linear range. However, the recovery from low-dilution samples was about 30% compared to almost 100% recovery for the high-dilution samples. The mean concentration value was also about three times higher with the high dilution. This clearly demonstrates the ion-suppression effects on CBG, and the elimination of this effect by sample dilution, as discussed above.[14] As CBG and CBD have similar retention times, the CBG signal was suppressed by CBD in high CBD samples. In the pool sample used in Table 3, the CBD concentration is >10 times that of CBG.
The method was applied to quantify cannabinoids in several different samples containing high levels of CBD and high levels of THC. The concentrations of each cannabinoid quantified are listed in Table 4.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Conclusions
An optimised, simple extraction method followed by a reliable and simple LC-MS method, was developed and validated for the simultaneous quantification of 17 cannabinoids. The method can be used by natural products laboratories that are equipped with a TOF-MS detector. By using low flow rate, the separation was achieved at low pressure (HPLC rather than UHPLC) conditions. The disadvantage of using the low flow rate was the long run time. Detection was performed by simpler MS rather than MS-MS. Although there was no significant loss of selectivity due to using MS instead of MS-MS, the sensitivity is compromised as evident from relatively high LOD and LOQ values (µg/mL levels compared to ng/mL levels commonly achieved with MS-MS). The main advantage of using this approach is that the method can be easily adapted to use with a simple HPLC-MS (single quadrupole) system that is commonly used in routine analytical laboratories. Due to the simplicity of instrumentation, and the robustness resulting from a high resolution in the chromatography of isobaric cannabinoids, the method is well suited for routine phytocannabinoid analysis across a range of applications in a variety of laboratory settings.
Acknowledgements
We want to thank Cann Group Limited, lead industry partner on the CRC-P, for their valuable input and for providing proprietary germplasm for this research.
Author contributions
Conceptualization A.K.H., T.K. and B.J.B.; methodology, A.K.H.; formal analysis A.K.H., F.G.-T. and M.N.; investigation, A.K.H., F.G.-T., S.P. and M.N.; writing—original draft preparation, A.K.H. and T.K.; writing—review and editing, B.J.B., F.G.-T., S.P. and M.N.; supervision, A.K.H. and T.K.; project administration, T.K. and S.P.; funding acquisition, B.J.B. and T.K. All authors have read and agreed to the published version of the manuscript.
Funding
This project received grant funding from the Australian Government via the Department of Industry, Science, Energy and Resources, as part of the Cooperative Research Centres Project (CRC-P) program—Round 7: Growing the medicinal cannabis industry—precision farming to pharmaceuticals.
Conflicts of interest
The authors declare no conflict of interest.
References
- ↑ Chandra, Suman; ElSohly, Mahmoud A.; Lata, Hemant, eds. (2017). Cannabis sativa L. - Botany and Biotechnology (1st ed. 2017 ed.). Cham: Springer International Publishing : Imprint: Springer. ISBN 978-3-319-54564-6.
- ↑ 2.0 2.1 Clarke, Robert C.; Merlin, Mark D. (1 November 2016). "Cannabis Domestication, Breeding History, Present-day Genetic Diversity, and Future Prospects" (in en). Critical Reviews in Plant Sciences 35 (5-6): 293–327. doi:10.1080/07352689.2016.1267498. ISSN 0735-2689. https://www.tandfonline.com/doi/full/10.1080/07352689.2016.1267498.
- ↑ 3.0 3.1 3.2 3.3 Giese, Matthew W; Lewis, Mark A; Giese, Laura; Smith, Kevin M (1 November 2015). "Method for the Analysis of Cannabinoids and Terpenes in Cannabis" (in en). Journal of AOAC INTERNATIONAL 98 (6): 1503–1522. doi:10.5740/jaoacint.15-116. ISSN 1060-3271. https://academic.oup.com/jaoac/article/98/6/1503-1522/5654599.
- ↑ van Breemen, Richard B.; Muchiri, Ruth N.; Bates, Timothy A.; Weinstein, Jules B.; Leier, Hans C.; Farley, Scotland; Tafesse, Fikadu G. (28 January 2022). "Cannabinoids Block Cellular Entry of SARS-CoV-2 and the Emerging Variants" (in en). Journal of Natural Products 85 (1): 176–184. doi:10.1021/acs.jnatprod.1c00946. ISSN 0163-3864. PMC PMC8768006. PMID 35007072. https://pubs.acs.org/doi/10.1021/acs.jnatprod.1c00946.
- ↑ Welling, Matthew T.; Liu, Lei; Hazekamp, Arno; Dowell, Ashley; King, Graham J. (25 February 2019). "Developing Robust Standardised Analytical Procedures for Cannabinoid Quantification: Laying the Foundations for an Emerging Cannabis-Based Pharmaceutical Industry" (in en). Medical Cannabis and Cannabinoids 2 (1): 1–13. doi:10.1159/000496868. ISSN 2504-3889. PMC PMC8489335. PMID 34676328. https://www.karger.com/Article/FullText/496868.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Citti, Cinzia; Braghiroli, Daniela; Vandelli, Maria Angela; Cannazza, Giuseppe (1 January 2018). "Pharmaceutical and biomedical analysis of cannabinoids: A critical review" (in en). Journal of Pharmaceutical and Biomedical Analysis 147: 565–579. doi:10.1016/j.jpba.2017.06.003. https://linkinghub.elsevier.com/retrieve/pii/S0731708517311895.
- ↑ 7.0 7.1 7.2 Micalizzi, Giuseppe; Vento, Federica; Alibrando, Filippo; Donnarumma, Danilo; Dugo, Paola; Mondello, Luigi (1 January 2021). "Cannabis Sativa L.: a comprehensive review on the analytical methodologies for cannabinoids and terpenes characterization" (in en). Journal of Chromatography A 1637: 461864. doi:10.1016/j.chroma.2020.461864. https://linkinghub.elsevier.com/retrieve/pii/S0021967320311389.
- ↑ 8.0 8.1 8.2 Hewavitharana, A.K.; Golding, G.; Tempany, G.; King, G.; Holling, N. (1 May 2005). "Quantitative GC-MS Analysis of Δ9-Tetrahydrocannabinol in Fiber Hemp Varieties" (in en). Journal of Analytical Toxicology 29 (4): 258–261. doi:10.1093/jat/29.4.258. ISSN 1945-2403. http://academic.oup.com/jat/article/29/4/258/730079/Quantitative-GCMS-Analysis-of.
- ↑ 9.0 9.1 ECA Foundation (8 April 2020). "German Pharmacopoeia Monograph for Cannabis Extracts". ECA Academy. ECA Foundation. https://www.gmp-compliance.org/gmp-news/german-pharmacopoeia-monograph-for-cannabis-extracts. Retrieved 28 February 2022.
- ↑ 10.0 10.1 Mudge, Elizabeth M.; Murch, Susan J.; Brown, Paula N. (1 May 2017). "Leaner and greener analysis of cannabinoids" (in en). Analytical and Bioanalytical Chemistry 409 (12): 3153–3163. doi:10.1007/s00216-017-0256-3. ISSN 1618-2642. PMC PMC5395585. PMID 28233028. http://link.springer.com/10.1007/s00216-017-0256-3.
- ↑ Berman, Paula; Futoran, Kate; Lewitus, Gil M.; Mukha, Dzmitry; Benami, Maya; Shlomi, Tomer; Meiri, David (1 December 2018). "A new ESI-LC/MS approach for comprehensive metabolic profiling of phytocannabinoids in Cannabis" (in en). Scientific Reports 8 (1): 14280. doi:10.1038/s41598-018-32651-4. ISSN 2045-2322. PMC PMC6155167. PMID 30250104. http://www.nature.com/articles/s41598-018-32651-4.
- ↑ Skoog, Douglas A. (1985). Principles of instrumental analysis. Saunders golden sunburst series (3rd ed.). Philadelphia: Saunders College Pub. ISBN 978-0-03-001229-7.
- ↑ Wang, Mei; Wang, Yan-Hong; Avula, Bharathi; Radwan, Mohamed M.; Wanas, Amira S.; van Antwerp, John; Parcher, Jon F.; ElSohly, Mahmoud A. et al. (1 December 2016). "Decarboxylation Study of Acidic Cannabinoids: A Novel Approach Using Ultra-High-Performance Supercritical Fluid Chromatography/Photodiode Array-Mass Spectrometry" (in en). Cannabis and Cannabinoid Research 1 (1): 262–271. doi:10.1089/can.2016.0020. ISSN 2378-8763. PMC PMC5549281. PMID 28861498. http://www.liebertpub.com/doi/10.1089/can.2016.0020.
- ↑ 14.0 14.1 14.2 Taylor, Paul J. (1 April 2005). "Matrix effects: the Achilles heel of quantitative high-performance liquid chromatography–electrospray–tandem mass spectrometry" (in en). Clinical Biochemistry 38 (4): 328–334. doi:10.1016/j.clinbiochem.2004.11.007. https://linkinghub.elsevier.com/retrieve/pii/S0009912004003364.
- ↑ 15.0 15.1 15.2 Hewavitharana, Amitha K. (1 January 2011). "Matrix matching in liquid chromatography–mass spectrometry with stable isotope labelled internal standards—Is it necessary?" (in en). Journal of Chromatography A 1218 (2): 359–361. doi:10.1016/j.chroma.2010.11.047. https://linkinghub.elsevier.com/retrieve/pii/S0021967310016213.
- ↑ 16.0 16.1 Hewavitharana, A.K.; Shawn, P.N.; Smyth, H.D.C. et al. (July 2021). "The Importance of Complete Overlapping of Analyte and Internal Standard Peaks in Eliminating Matrix Effects with Liquid Chromatography–Mass Spectrometry (LC–MS)". LCGC North America 39 (7): 335–38. https://www.chromatographyonline.com/view/the-importance-of-complete-overlapping-of-analyte-and-internal-standard-peaks-in-eliminating-matrix-effects-with-liquid-chromatography-mass-spectrometry-lc-ms-.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added.