Difference between revisions of "Journal:Design and evaluation of a LIS-based autoverification system for coagulation assays in a core clinical laboratory"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 18: | Line 18: | ||
|website = [https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-019-0848-2 https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-019-0848-2] | |website = [https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-019-0848-2 https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-019-0848-2] | ||
|download = [https://bmcmedinformdecismak.biomedcentral.com/track/pdf/10.1186/s12911-019-0848-2 https://bmcmedinformdecismak.biomedcentral.com/track/pdf/10.1186/s12911-019-0848-2] (PDF) | |download = [https://bmcmedinformdecismak.biomedcentral.com/track/pdf/10.1186/s12911-019-0848-2 https://bmcmedinformdecismak.biomedcentral.com/track/pdf/10.1186/s12911-019-0848-2] (PDF) | ||
}} | }} | ||
| Line 91: | Line 85: | ||
===Assessment of the validation of the autoverification system=== | ===Assessment of the validation of the autoverification system=== | ||
====Assessment of the historical records test | ====Assessment of the historical records test==== | ||
According to CLSI's AUTO 10-A guidance, all rules and settings were required to be validated and thoroughly tested before the autoverification protocols were implemented in a real-life clinical environment. First, we used historical clinical laboratory records stored in the LIS to verify the validation. To test if the program would screen the results outside the autoverification decision rules, special abnormal cases were chosen to test whether or not the autoverification program could be held up. Second, the autoverification passing rate of historical records in the LIS from January 2017 to March 2017 was determined to test if the autoverification protocols followed the rules as expected and to detect potential problems with the program. | According to CLSI's AUTO 10-A guidance, all rules and settings were required to be validated and thoroughly tested before the autoverification protocols were implemented in a real-life clinical environment. First, we used historical clinical laboratory records stored in the LIS to verify the validation. To test if the program would screen the results outside the autoverification decision rules, special abnormal cases were chosen to test whether or not the autoverification program could be held up. Second, the autoverification passing rate of historical records in the LIS from January 2017 to March 2017 was determined to test if the autoverification protocols followed the rules as expected and to detect potential problems with the program. | ||
| Line 451: | Line 445: | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Randell ''et al.''<ref name="RandellStrat18" /> | | style="background-color:white; padding-left:10px; padding-right:10px;"|Randell ''et al.''<ref name="RandellStrat18" /> | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|2018 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Canada | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Clinical Chemistry and immunoassay | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Middleware | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|80,867 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|91.1 to 94.7% | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Xia ''et al.''<ref name="XiaDevelop17">{{cite journal |title=Developing and application of an autoverification system for clinical chemistry and immunology test results |journal=National Medical Journal of China |author=Xia, L.Y.; Cheng, X.Q.; Liu, Q. et al. |volume=97 |issue=8 |pages=616-621 |year=2017 |doi=10.3760/cma.j.issn.0376-2491.2017.08.012 |pmid=28260308}}</ref> | | style="background-color:white; padding-left:10px; padding-right:10px;"|Xia ''et al.''<ref name="XiaDevelop17">{{cite journal |title=Developing and application of an autoverification system for clinical chemistry and immunology test results |journal=National Medical Journal of China |author=Xia, L.Y.; Cheng, X.Q.; Liu, Q. et al. |volume=97 |issue=8 |pages=616-621 |year=2017 |doi=10.3760/cma.j.issn.0376-2491.2017.08.012 |pmid=28260308}}</ref> | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|2017 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|China | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Clinical Chemistry and immunoassay | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|LIS | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|31,349 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|74.0% | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Krasowski ''et al.''<ref name="KrasowkskiAuto14">{{cite journal |title=Autoverification in a core clinical chemistry laboratory at an academic medical center |journal=Journal of Pathology Informatics |author=Krasowski, M.D.; Davis, S.R.; Drees, D. et al. |volume=5 |issue=1 |page=13 |year=2014 |doi=10.4103/2153-3539.129450 |pmid=24843824 |pmc=PMC4023033}}</ref> | | style="background-color:white; padding-left:10px; padding-right:10px;"|Krasowski ''et al.''<ref name="KrasowkskiAuto14">{{cite journal |title=Autoverification in a core clinical chemistry laboratory at an academic medical center |journal=Journal of Pathology Informatics |author=Krasowski, M.D.; Davis, S.R.; Drees, D. et al. |volume=5 |issue=1 |page=13 |year=2014 |doi=10.4103/2153-3539.129450 |pmid=24843824 |pmc=PMC4023033}}</ref> | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|2014 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|United States | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Clinical Chemistry and immunoassay | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Middleware | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|99.5% | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Palmieri ''et al.''<ref name="PalmieriTheDev18">{{cite journal |title=The development of autoverification rules applied to urinalysis performed on the AutionMAX-SediMAX platform |journal=Clinical Chimica Acta |author=Palmieri, R.; Falbo, R.; Cappellini, F. et al. |volume=485 |pages=275–81 |year=2018 |doi=10.1016/j.cca.2018.07.001 |pmid=29981288}}</ref> | | style="background-color:white; padding-left:10px; padding-right:10px;"|Palmieri ''et al.''<ref name="PalmieriTheDev18">{{cite journal |title=The development of autoverification rules applied to urinalysis performed on the AutionMAX-SediMAX platform |journal=Clinical Chimica Acta |author=Palmieri, R.; Falbo, R.; Cappellini, F. et al. |volume=485 |pages=275–81 |year=2018 |doi=10.1016/j.cca.2018.07.001 |pmid=29981288}}</ref> | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|2018 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Italy | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Urinalysis | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|AutionMAX-SediMAX | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|1,002 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|52.4% | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Sediq ''et al.''<ref name="SediqDesign14" /> | | style="background-color:white; padding-left:10px; padding-right:10px;"|Sediq ''et al.''<ref name="SediqDesign14" /> | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|2014 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Egypt | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Thyroid Function | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|LIS | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|563 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|63.8% | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Zhao ''et al.''<ref name="ZhaoBuilding14" /> | | style="background-color:white; padding-left:10px; padding-right:10px;"|Zhao ''et al.''<ref name="ZhaoBuilding14" /> | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|2014 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|China | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Coagulation | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Middleware | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|2,353 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|82.0% | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Our study | | style="background-color:white; padding-left:10px; padding-right:10px;"|Our study | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|China | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|Coagulation | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|LIS | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|83,699 | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | | style="background-color:white; padding-left:10px; padding-right:10px;"|78.86% | ||
|- | |- | ||
|} | |} | ||
|} | |} | ||
====Efficiency of the autoverification system==== | |||
Comparing the TAT from July to December 2017 with that from July to December 2016, for the routine coagulation assay samples, the median of TAT 1 was 126 minutes before using the autoverification system versus 101 minutes after processing the autoverification system. The median of TAT 2 was shortened from 41 minutes to 15 minutes. Statistically significant differences were observed for TAT 1 and TAT 2 (P < 0.001, P < 0.001, respectively, Mann-Whitney U test). | |||
==Discussion== | |||
Coagulation profiles were chosen for autoverification because coagulation provides essential and timely analytical information and is crucial for patient treatment and evaluation of physical condition. In our hospital, the numbers of routine coagulation profiles—including PT, APTT, TT and FBG—exceeded 12,000 routine samples per month, and autoverification could be the appropriate answer to process the huge workloads and save labor. Until recently, laboratory-based autoverification that integrates into LIS was still relatively rare. | |||
Our study has taken all factors into consideration, including the instruments, standard serum samples, reagents, specimens, and the logical relationship of the test items. We designed an autoverification system based on a set of rules integrated with the LIS that runs in the following order: QC check, analytical error flags, critical value, limited range check, delta check, logical rules, and clinical diagnosis, as shown previously in Fig. 1. If any of the rules are violated, the outcome will be held up in the LIS for manual verification. QC was the premise of the autoverification, and analytical error flags and critical value were performed as the initial screening to avoid basic analytical error. | |||
The key points of the autoverification protocols are building the limited range check, delta check, and logical check. The limited range check is vital to samples without historical data, because for those patients the delta check is not available. In our laboratory, because the LIS is connected with the HIS, the patient’s diagnosis would be considered first, e.g., if they were undergoing Warfarin treatment, the limited range was PT > 40.00 s, and the allowable range for INR was changed. Thus, a limited range check that corresponds with patient’s clinical diagnosis or health conditions facilitates the possibility of precise and customized autoverification. A delta check facilitated the technologists to quickly recognize changes in a patient’s status. Patient’s results with historical data failing the limited range but passing the delta check would pass the autoverification. In this circumstance, our LIS-based autoverification can potentially provide verification more precisely than commercial tools or manual verification. Logical rules were applied to enable the technologists to quickly detect the logical relationship between the items and analytical errors. | |||
Our research extends the knowledge into LIS-based autoverification. We defined the limited range based on the large-scale data of 157,079 routine coagulation assays for one year, and we observed the validation and efficiency of autoverification protocols based on 83,699 samples for half a year to report the validation and benefits of autoverification. The validation of the autoverification rules was assessed through both the historical data and online test version program. When using the historical data for analysis, the autoverification passing rate was 77.28%. During the online test version program, the passing rate was 77.63% in May 2017 and 78.75% in June 2017. We modified the logical rules to minimize the false-negative rate because we believe that the crucial point of autoverification is to guarantee the best medical safety. The average passing rate was 78.86%, and the 95% CI for the overall passing rate was [78.25, 79.59%] when we formally implemented the program. Our results suggest that the decision rules permit the autoverification system to achieve better accuracy and guarantee medical safety with the availability of historical data and diagnosis. | |||
To date, as shown in Table 5, the passing rate of the autoverification system is similar to the results based on other commercial tools or middleware<ref name="ZhaoBuilding14" /><ref name="PalmieriTheDev18" />, and the passing rate for coagulation in our laboratory is to some extent higher than those of other studies that use autoverification integrated with LIS.<ref name="SediqDesign14" /><ref name="XiaDevelop17" /> Interestingly, Randell ''et al.''<ref name="RandellStrat18" /><ref name="RandellAuto18" /> and Krasowski ''et al.''<ref name="KrasowkskiAuto14" /> reported higher autoverification passing rates of more than 90%. Well-designed limit range sets and the selective use of delta checks may contribute to the high autoverification passing rates in their research.<ref name="RandellStrat18" /><ref name="RandellAuto18" /> Both studies focused on clinical chemistry and immunoassay tests, whereas our study focused on coagulation. It is generally accepted that clinical chemistry and immunoassay are common and regular tests for outpatients, inpatients, and patients from physical examination centers, while in contrast, coagulation is to some extent specifically prescribed for sick patients in emergencies, undergoing anticoagulant or thrombolytic therapy, or for patients in pregnancy. Therefore, pre-analytical problems and problematic samples could be the reason for the lower passing rate of coagulation. In addition, our hospital specifically treats perplexing and complicated illnesses in Northeast China, and many samples from severely ill patients were collected, which may enhance the proportion of problematic samples in coagulation assays. | |||
Compared to other studies that assessed the inter-observer degree of agreement between manual verification and autoverification<ref name="LiDesign18" /><ref name="SediqDesign14" />, we analyzed the true-positive rate, the true-negative rate, the false-positive rate, the false-negative rate, the sensitivity, and the specificity. In our study, no false-negative case was observed prior to the high passing rate to ensure that every sample that passes the autoverification system and is auto-released is correctly reported, thus guaranteeing a high level of medical safety. This is because higher autoverification passing rates and lower false-positive rates may inevitably lead to higher false-negative rates, and thus some problematic samples would be released without attentive manual verification or re-analysis. | |||
When determining the efficiency of autoverification, TAT is the indication most affected by the implementation of autoverification.<ref name="TorkeProcess05" /> In the present study, the TAT 1 and TAT 2 were shortened, which is consistent with previous reports.<ref name="SediqDesign14" /><ref name="ZhaoBuilding14" /> Because the passing autoverification are released immediately to HIS, the TAT is significantly reduced compared with those processed by manual verification, which in turn could save time and decrease the manual workload in clinical laboratories.<ref name="RandellStrat18" /> Furthermore, another benefit is that the autoverification system follows consistent standardized processes, therefore minimizing the potential mistakes made by laboratory technologists due to lack of clinical experience or feelings of fatigue and stress. | |||
This study has limitations to be considered. First, it is difficult to assess the error rate of the autoverification system. Because no obvious mistakes or errors have been reported by patients or clinicians at our hospital for nearly two years, we may infer that the autoverification protocols are at least as safe as the previous manual verification. Second, a higher passing rate of the autoverification system should be designed with future research. | |||
==Conclusions== | |||
We developed and assessed an autoverification system for the most commonly used coagulation profile with a set of rules. The autoverification system for coagulation assays based on LIS can halt the samples with abnormal values for manual verification, guarantee medical safety, minimize the requirements for manual work, shorten TAT, and raise working efficiency. Furthermore, we desire to design more precise and valuable rules to build a model in which autoverification may best be used on coagulation profiles. | |||
==Availability of data and materials== | |||
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. | |||
==Abbreviations== | |||
'''APTT''': Activated partial thromboplastin time | |||
'''AUTO 10-A''': ''Autoverification of Clinical Laboratory Test Results'' by CLSI | |||
'''FBG''': Fibrinogen | |||
'''HIS''': Hospital information system | |||
'''INR''': International normalized ratio | |||
'''LIS''': Laboratory information system | |||
'''PT''': Prothrombin time | |||
'''TAT''': Turnaround time | |||
'''TT''': Thrombin time | |||
==Acknowledgements== | |||
===Funding=== | |||
This research was supported by the Humanities Social Science Foundation of Ministry of Education (Program Grant 17YJCZH184) and Liaoning Education Science 13th Five-Year plan (Program Grant JG17DB557); Philosophy and Social Sciences promotion project of China Medical University (Program Grant 111–3110118083); and The National Natural Science Foundation (Program Grant 81700814). The funding bodies were not involved in neither the design of the study nor the collection, analysis, and interpretation of data, nor the writing of the manuscript. | |||
===Author contributions=== | |||
ZW, HK, and BQ designed the study. ZW, HK, XF, RM, LZ, and MH undertook data collection. CP and BQ analyzed the data. ZW and CP wrote drafts of the manuscript. Then, BQ revised drafts of the paper. All authors contributed to the interpretation of findings and have read and approved the final manuscript. | |||
===Ethics declarations=== | |||
Ethics approval was obtained from the Ethics Committee of the First Affiliated Hospital of China Medical University (2015<ref name="StrathmannSim11" />). The patient consent was waived, as utilization of anonymized history data does not require patient consent. | |||
===Competing interests=== | |||
The authors declare that they have no competing interests. | |||
==References== | ==References== | ||
Latest revision as of 23:47, 1 October 2019
| Full article title | Design and evaluation of a LIS-based autoverification system for coagulation assays in a core clinical laboratory |
|---|---|
| Journal | BMC Medical Informatics and Decision Making |
| Author(s) | Wang, Zhongqing; Peng, Cheng; Kang, Hui; Fan, Xia; Mu, Runqing; Zhou, Liping, He, Miao; Qu, Bo |
| Author affiliation(s) | China Medical University, Affiliated Hospitals of China Medical University |
| Primary contact | Online contact form |
| Year published | 2019 |
| Volume and issue | 19(1) |
| Page(s) | 123 |
| DOI | 10.1186/s12911-019-0848-2 |
| ISSN | 1472-6947 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-019-0848-2 |
| Download | https://bmcmedinformdecismak.biomedcentral.com/track/pdf/10.1186/s12911-019-0848-2 (PDF) |
Abstract
Background: An autoverification system for coagulation consists of a series of rules that allows normal data to be released without manual verification. With new advances in medical informatics, the laboratory information system (LIS) has growing potential for the use of autoverification, allowing rapid and accurate verification of clinical laboratory tests. The purpose of the study is to develop and evaluate a LIS-based autoverification system for validation and efficiency.
Methods: Autoverification decision rules—including quality control, analytical error flag, critical value, limited range check, delta check, and logical check rules, as well as patient’s historical information—were integrated into the LIS. Autoverification limit ranges was constructed based on 5% and 95% percentiles. The four most commonly used coagulation assays—prothrombin time (PT), activated partial thromboplastin time (APTT), thrombin time (TT), and fibrinogen (FBG)—were followed by the autoverification protocols. The validation was assessed using characteristics such as autoverification passing rate, the true-positive cases, the true-negative cases, the false-positive cases, the false-negative cases, the sensitivity and the specificity. Efficiency was evaluated by turnaround time (TAT).
Results: A total of 157,079 historical test results of coagulation profiles from January 2016 to December 2016 were collected to determine the distribution intervals. The autoverification passing rate was 77.11% (29,165 / 37,821) based on historical patient data. In the initial test of the autoverification version in June 2017, the overall autoverification passing rate for the whole sample was 78.75% (11,257 / 14,295), with 892 true-positive cases, 11,257 true-negative cases, 2,146 false-positive cases, no false-negative cases, sensitivity of 100% ,and specificity of 83.99%. After formal implementation of the autoverification system for six months, 83,699 samples were assessed. The average overall autoverification passing rate for the whole sample was 78.86%, and the 95% confidence interval (CI) of the passing rate was [78.25, 79.59%]. TAT was reduced from 126 minutes to 101 minutes, which was statistically significant (P < 0.001, Mann-Whitney U test).
Conclusions: The LIS-based autoverification system for coagulation assays can halt the samples with abnormal values for manual verification, guarantee medical safety, minimize the requirements for manual work, shorten TAT, and raise working efficiency.
Keywords: laboratory information systems, medical safety, autoverification, coagulation, turnaround time
Background
Following the analytical phase, a large number of manual verifications are performed in clinical laboratories to detect possible errors before results are released to electronic health records (EHRs), which is time-consuming.[1] A significant solution for this issue may be autoverification, a process which uses a set of well-designed rules to identify and flag samples with abnormal values for manual verification, at the same time permitting those with normal values to be released without manual intervention.[2] Previous reports have demonstrated that autoverification can ensure medical safety[3], shorten turnaround time (TAT)[2][3][4][5], reduce labor requirements[2][3][4], improve operational efficiency[2][4][5][6] and minimize error rate[2], as well as enable laboratory technologists to devote more attention to test results that have greater potential for error.[2]
Until recently, those autoverification systems were commonly developed via third-party commercial software or middleware, which are costly, and the autoverification decision rules were proprietary, such that no revision could be made according to user requirements.[7][8][9][10] In addition, they could not connect with a hospital information system (HIS) and obtain comprehensive clinical data, such as a patient’s history and clinical diagnosis. With the rapid progress of laboratory automation today, it is challenging to achieve interconnection and intercommunication between analytical instruments and the laboratory information system (LIS) with the goal of designing laboratory-focused autoverification systems independent of any commercial software.
Coagulation assays are essential for the assessment of patients requiring acute care[11], patients undergoing anticoagulant therapy[12], thrombolytic therapy[13], and pregnancy[14], as well as for the monitoring of disseminated intravascular coagulation.[15] In many laboratories, coagulation assays are currently still released by manual review, verification, and release, and reports about autoverification in coagulation are scarce.[16] The four most routinely used coagulation assays in our laboratory—namely, prothrombin time (PT), activated partial thromboplastin time (APTT), thrombin time (TT), and fibrinogen (FBG)—are commonly prescribed together. As such, an urgent need to automate these assays exists, yet it is still a challenge to establish autoverification decision rules in coagulation that take advantage of quality control (QC) checks, instrument error flags, critical value warnings, limited range checks, delta checks, logical rule requirements, and patients' historical data, all while referencing the Clinical Laboratory Standards Institute (CLSI) guidelines for Autoverification of Clinical Laboratory Test Results (AUTO 10-A).[17] This research extends our knowledge to establish a laboratory-specific autoverification system for coagulation assays based on LIS to ensure medical safety and shorten turnaround time (TAT). Moreover, it is the first study based on clinical large-scale data to evaluate the validation and efficiency of autoverification in terms of specificity and efficiency.
Methods
Setting
The study was conducted in the core clinical laboratory of the First Affiliated Hospital of China Medical University, which processes approximately 3,500,000 various laboratory samples per year. The HIS and the LIS were both provided by Beijing Donghua Software (Beijing, Donghua, China). The autoverification decision rules were converted into computer languages to integrate into the HIS. In the process of autoverification, the values that passed the autoverification protocols were marked in green, and those that failed were flagged in red. When abnormal results were held up, a quick attentive manual verification or re-analysis was needed. If the results were considered to be critical values, they were quickly released to the HIS, and the clinicians were alerted to the abnormalities by a call.
Data collection
Coagulation profiles from outpatients, inpatients, and patients from the physical examination center of our hospital were collected for analysis. Historical values of 157,097 samples between January 2016 and December 2016 were used to determine the 5% and 95% percentiles of PT, APTT, TT and FBG. A total of 37,821 historical records released by manual verification from January 2017 to March 2017 were obtained to test the passing rate of autoverification protocols. In the test version phase, validation of the autoverification protocols was determined by 28,244 samples between May 2017 and June 2017. After the protocols were formally implemented online, 83,699 samples between July 2017 and December 2017 were taken to evaluate the single test autoverification passing rate and the overall autoverification passing rate.
Analytical instruments and reagents
All tests were measured using STA-R Evolution Analyzer (Stago Diagnostica, Brussels, Belgium). Reagents of STA-PTT (APTT), STA-Neoplastin R (PT), STA-TT (TT), STA-Fibrinogen (FBG), and controls were also provided by Stago.
Autoverification decision making
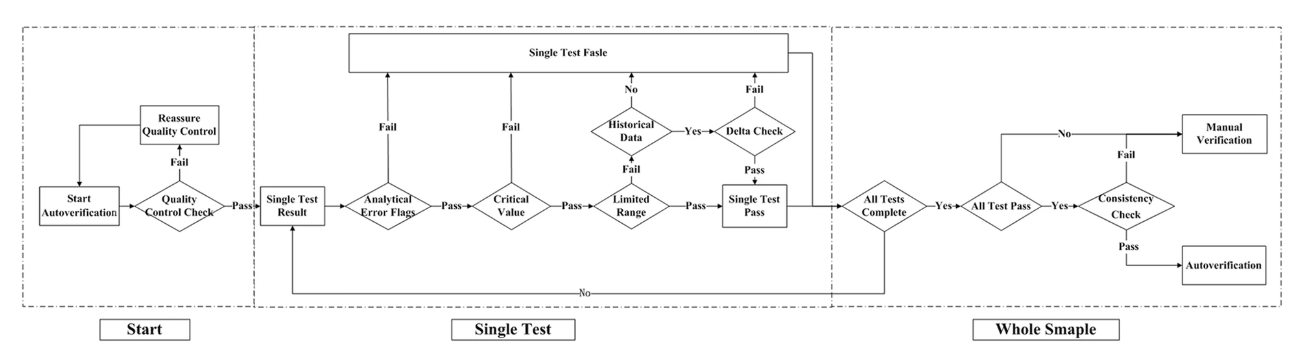
The autoverification system was designed with the (CLSI) guidelines for Autoverification of Clinical Laboratory Test Results (AUTO 10-A) in mind.[17] A technologist-augmented decision-making tree was processed, and if any of the rules were violated, the result was held for manual verification, as shown in Fig. 1. The autoverification steps consisted of quality control (QC) checks, analytical error flags, critical values, limited range checks, delta checks, and logical rules, as well as confirming the patient’s clinical diagnosis. The autoverification protocols ran the single test successively until the four items of the whole sample were finished. If all four items passed the autoverification, the logical rules were run for further verification; if any of the steps violated the rules, manual verification was required. Because the LIS is connected to the HIS in our hospital, any result that passed autoverification was directly released to the patients’ medical records without manual verification.
|
Quality control check
A quality control (QC) check is routinely performed in our laboratory, transmitting from the analyzer to the LIS according to Westgard quality control multi-rules.[18] The QC check ensured that each test was within the appropriate range before running the autoverification protocols. If there were no QC results within 24 hours or if any QC rule was violated, the program halted all samples being autoverified.
Analytical error flags
If any instrument analytical flag was sent to the LIS along with testing problems, such as the two-dimensional barcode, reagents, and samples, as well as reagent crystallization or sample clots forming, the results were held for later manual verification.
Critical value
The critical values that may threaten the patient’s life and require urgent medical treatment were determined by laboratory technologists and clinicians. The critical values used for autoverification were as follows: PT ≤ 9.00 s or ≥ 70.00 s, APTT ≤ 15.00 s or ≥ 100.00 s, TT > 150.00 s and FBG < 1.00 g/L (as shown later on in Table 2). If critical values were detected, they were manually verified by a skilled laboratory specialist, repeated, and transferred to a critical value list for an immediate phone call to clinicians. The results found to be within the range proceeded to the next steps: limited range check and delta check.
Limited range check
The limited range is a filter to verify if the result is within the extreme reference intervals specified for the assays in the LIS. The clinical reference intervals used in our laboratory were 11.00–14.30 s for PT, 32.00–43.00 s for APTT, 14.00–21.00 s for TT, and 2.00–4.00 g/L for FBG. In clinical practice, PT and TT values greater than 3.00 s and APTT values greater than 100.00 s are considered pathological. However, due to biological variation, clinical reference intervals are not always suitable for autoverification; we therefore analyzed the distribution of 5% and 95% percentiles based on the historical large-scale data from January 2016 to December 2016. In the current study, after discussing with local clinicians, the limited range was determined by a combination of clinical reference intervals, critical value, and 5% and 95% percentiles. The results that were within the limited range continued to the next step.
Delta check
It is widely accepted that test results with historical values should be confirmed by a delta check.[19][20] In our study, delta check was used for the results with historical data and failing the limited check. The delta check was performed to compare present test results with previous results and to identify variations beyond a patient's expected values, such as changes in a patient's clinical condition or specimens with clots present.[20][21][22][23][24] Delta check can be performed using various methods, including the delta difference, delta percent change, rate difference, and rate percent difference.[22][25][26] In the present study, delta differences were calculated as the difference between the value of the present results and the previous results. Meanwhile, different tests correspond to different delta differences and time intervals due to biological and pathological functions of coagulation assays. The delta check rules were as follows: PT < 10.00 s and the time interval < 10 days; APTT < 10.00 s and the time interval < 7 days; FBG < 1.00 g/L and the time interval < 3 days. However, if the patient was undergoing anticoagulant therapy, the time interval was < 30 days, and the international normalized ratio (INR) was between 2 and 3.
Logical rules
Based on clinical and practical diagnostic criteria, a few tests may have some necessary logical interrelationships between each testing item, e.g., prolonged PT and APTT associated with a decreased FBG. Logical rules were applied with the condition that all single tests passed the QC check, analytical error flags, critical value check, limited range check, and delta check. If any result violated the logical rules, the autoverification protocols were stopped and the results were reviewed manually before being sent to the HIS. During the logical check, even though the single results were within the limited range, the reports were held for manual verification when the following occurred: PT ≥ 14.00 s or APTT ≥ 43.00 s and FBG ≤ 2.00 g/L; PT ≥ 14.00 s and APTT ≥ 43.00 s; or APTT ≤ 43.00 s, PT ≤ 14.00 s, and TT ≥ 21.00 s. By logical rules, a presence of clot was avoided.
Patient’s clinical diagnosis and historical data
Because it is of great significance for us to design precise and customized rules that work for inpatients and outpatients, patient-specific information—such as gender, age, clinical presentation, medical treatment history, drug history, pregnancy, and other conditions that may affect the results of coagulation assays—should be comprehensively considered. Adjustment of the limited range was done according to clinical diagnosis and historical data. For instance, for patients taking Warfarin, the limited range was PT > 40.00 s, and the allowable ranges for INR were as follows: 1.50–2.50 before non-hip surgery, 2.00–3.00 before hip surgery, 2.00–3.00 for deep vein thrombosis, 2.00–4.00 for treatment of pulmonary infarction, 3.00–4.00 for prevention of arterial thrombosis, and 3.00–4.00 for valve prosthesis surgery. For patients receiving heparin treatment, the limited range was APPT > 90.00 s. If the patient was undergoing anticoagulation therapy, even though the test result of PT was within the routine limited range rather than within the customized limited range, the report would be stopped for manual autoverification and would not be sent to the HIS. For late pregnancy, the upper limit intervals for PT and APTT were decreased by 10%, and the lower limited range for FBG was increased by 10%.[14] For patients undergoing thrombolytic therapy, the limited range for FBG was defined as 1.20–4.00 g/L.
Assessment of the validation of the autoverification system
Assessment of the historical records test
According to CLSI's AUTO 10-A guidance, all rules and settings were required to be validated and thoroughly tested before the autoverification protocols were implemented in a real-life clinical environment. First, we used historical clinical laboratory records stored in the LIS to verify the validation. To test if the program would screen the results outside the autoverification decision rules, special abnormal cases were chosen to test whether or not the autoverification program could be held up. Second, the autoverification passing rate of historical records in the LIS from January 2017 to March 2017 was determined to test if the autoverification protocols followed the rules as expected and to detect potential problems with the program.
Assessment of the initial online test version and autoverification system
Subsequently, the initial online test version program was run in a real-life environment to verify the actual validation in May 2017. In the test version phase, the autoverification decision rules were programmed in other computers, and the autoverification reports were not sent to the HIS. Meanwhile, a laboratory technologist and a skilled laboratory specialist (professor in the core clinical laboratory) manually verified and revised all routine cases and then released reports to HIS as before. Comparisons were accomplished as follows: the number of samples, cases that passed the autoverification, cases that failed the autoverification, the true-positive cases (intercepted problematic reports), the true-negative cases (auto-released correct reports), the false-positive cases (intercepted correct problematic reports), the false-negative cases (auto-released problematic reports), the sensitivity, the specificity, and the autoverification passing rate. After initial online testing for one month, false-negative cases were found due to a tiny or partial clot that was not detected by the instrument. Thus, logical rules were modified to eliminate the false-negative rate in June 2017. Finally, after running and modifying the test version, the false-negative rate was zero, and the program was formally connected online with the LIS in July 2017. The results of all of the samples that passed all the autoverification decision rules were then directly released from the LIS to the HIS without manual verification.
Assessment of the efficiency of the autoverification system
In the current study, TAT was measured by two means, namely TAT 1 and TAT 2. TAT 1 is defined as the interval from the sample receipt by the laboratory to the release of the results on the LIS, while TAT 2 is defined as the interval from the complete analysis by the instruments to the report verification on LIS. Both TAT 1 and TAT 2 were compared with the corresponding previous months.
Statistical analysis
Data were analyzed using IBM SPSS statistics software, version 23.0 (SPSS Inc., Chicago, USA). The Mann-Whitney U test was used for independent sample analysis. A P < 0.05 was considered significant.
Results
Definition of limited range
From January 2016 and December 2016, 157,079 historical test results of coagulation profiles were collected. Distribution intervals of clinical large-scale data of coagulation, including the mean, standard deviation, median, and 5% and 95% percentiles, are shown in Table 1.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
In this study, we defined the limited range according the intervals of clinical reference intervals, critical range, and 5% and 95% percentiles, and the limited range was 11.00–16.30 s for PT, 30.40–46.40 s for APTT, 14.00–21.00 s for TT, and 2.00–6.51 g/L for FBG, as shown in Table 2.
| ||||||||||||||||||||||||||||||
Validation of the autoverification system
Validation of historical records
In historical data analysis, we evaluated 246 items of historical special abnormal data with high or low concentrations. After running through the autoverification protocols and comparing with the historical reports, all items ended in stopping autoverification and none ended in autoverification. Of the 37,821 historical reports of coagulation profiles from January 2017 to March 2017, 29,165 samples passed the autoverification protocols, indicating that the overall passing rate was 77.11%.
Validation of initial online test
The autoverification passing rate, number of samples, cases that passed the autoverification, cases that failed the autoverification, true-positive cases, true-negative cases, false-positive cases, false-negative cases, sensitivity, and specificity in the real-life online test are shown in Table 3. Because coagulation requisition forms in our hospital usually contained PT, APTT, FBG, and TT simultaneously, we evaluated the overall passing rate of the four tests as a whole sample. The overall autoverification passing rate for all of the samples was 77.63% (10,828/13,949) in May 2017. There were five cases that the two reviewers decided to stop for manual verification; however, the autoverification passed them. These cases were considered to be instances of a partial or tiny clot, characterized by PT ≥ 14.00 s or APTT ≥ 43.00 s and FBG ≤ 2.00 g/L, despite being within limited range. After modifying the protocols with logical rules, no false-negative cases occurred, and the passing rate was 78.75% (11,257/14,295) in June 2017.
| ||||||||||||||||||||||||||||||||||||
Validation of the autoverification system
In July 2017, autoverification was formally introduced, and we observed 83,699 samples from July 2017 to December 2017 to assess the validation. No cases failed the QC check, and approximately 120 items failed analytical error flags per month. Rules of critical value, limited range, and delta check were determined by single tests, and the autoverification passing rate is shown in Table 4.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
After modifying the logical rules, the average overall passing rate was 78.86%, and the 95% confidence interval (CI) for the overall passing rate was [78.25, 79.59%]. To compare with other studies, the passing rates of different assays and systems (commercial tools, middleware, or LIS) are shown in Table 5.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Efficiency of the autoverification system
Comparing the TAT from July to December 2017 with that from July to December 2016, for the routine coagulation assay samples, the median of TAT 1 was 126 minutes before using the autoverification system versus 101 minutes after processing the autoverification system. The median of TAT 2 was shortened from 41 minutes to 15 minutes. Statistically significant differences were observed for TAT 1 and TAT 2 (P < 0.001, P < 0.001, respectively, Mann-Whitney U test).
Discussion
Coagulation profiles were chosen for autoverification because coagulation provides essential and timely analytical information and is crucial for patient treatment and evaluation of physical condition. In our hospital, the numbers of routine coagulation profiles—including PT, APTT, TT and FBG—exceeded 12,000 routine samples per month, and autoverification could be the appropriate answer to process the huge workloads and save labor. Until recently, laboratory-based autoverification that integrates into LIS was still relatively rare.
Our study has taken all factors into consideration, including the instruments, standard serum samples, reagents, specimens, and the logical relationship of the test items. We designed an autoverification system based on a set of rules integrated with the LIS that runs in the following order: QC check, analytical error flags, critical value, limited range check, delta check, logical rules, and clinical diagnosis, as shown previously in Fig. 1. If any of the rules are violated, the outcome will be held up in the LIS for manual verification. QC was the premise of the autoverification, and analytical error flags and critical value were performed as the initial screening to avoid basic analytical error.
The key points of the autoverification protocols are building the limited range check, delta check, and logical check. The limited range check is vital to samples without historical data, because for those patients the delta check is not available. In our laboratory, because the LIS is connected with the HIS, the patient’s diagnosis would be considered first, e.g., if they were undergoing Warfarin treatment, the limited range was PT > 40.00 s, and the allowable range for INR was changed. Thus, a limited range check that corresponds with patient’s clinical diagnosis or health conditions facilitates the possibility of precise and customized autoverification. A delta check facilitated the technologists to quickly recognize changes in a patient’s status. Patient’s results with historical data failing the limited range but passing the delta check would pass the autoverification. In this circumstance, our LIS-based autoverification can potentially provide verification more precisely than commercial tools or manual verification. Logical rules were applied to enable the technologists to quickly detect the logical relationship between the items and analytical errors.
Our research extends the knowledge into LIS-based autoverification. We defined the limited range based on the large-scale data of 157,079 routine coagulation assays for one year, and we observed the validation and efficiency of autoverification protocols based on 83,699 samples for half a year to report the validation and benefits of autoverification. The validation of the autoverification rules was assessed through both the historical data and online test version program. When using the historical data for analysis, the autoverification passing rate was 77.28%. During the online test version program, the passing rate was 77.63% in May 2017 and 78.75% in June 2017. We modified the logical rules to minimize the false-negative rate because we believe that the crucial point of autoverification is to guarantee the best medical safety. The average passing rate was 78.86%, and the 95% CI for the overall passing rate was [78.25, 79.59%] when we formally implemented the program. Our results suggest that the decision rules permit the autoverification system to achieve better accuracy and guarantee medical safety with the availability of historical data and diagnosis.
To date, as shown in Table 5, the passing rate of the autoverification system is similar to the results based on other commercial tools or middleware[16][29], and the passing rate for coagulation in our laboratory is to some extent higher than those of other studies that use autoverification integrated with LIS.[5][27] Interestingly, Randell et al.[6][8] and Krasowski et al.[28] reported higher autoverification passing rates of more than 90%. Well-designed limit range sets and the selective use of delta checks may contribute to the high autoverification passing rates in their research.[6][8] Both studies focused on clinical chemistry and immunoassay tests, whereas our study focused on coagulation. It is generally accepted that clinical chemistry and immunoassay are common and regular tests for outpatients, inpatients, and patients from physical examination centers, while in contrast, coagulation is to some extent specifically prescribed for sick patients in emergencies, undergoing anticoagulant or thrombolytic therapy, or for patients in pregnancy. Therefore, pre-analytical problems and problematic samples could be the reason for the lower passing rate of coagulation. In addition, our hospital specifically treats perplexing and complicated illnesses in Northeast China, and many samples from severely ill patients were collected, which may enhance the proportion of problematic samples in coagulation assays.
Compared to other studies that assessed the inter-observer degree of agreement between manual verification and autoverification[4][5], we analyzed the true-positive rate, the true-negative rate, the false-positive rate, the false-negative rate, the sensitivity, and the specificity. In our study, no false-negative case was observed prior to the high passing rate to ensure that every sample that passes the autoverification system and is auto-released is correctly reported, thus guaranteeing a high level of medical safety. This is because higher autoverification passing rates and lower false-positive rates may inevitably lead to higher false-negative rates, and thus some problematic samples would be released without attentive manual verification or re-analysis.
When determining the efficiency of autoverification, TAT is the indication most affected by the implementation of autoverification.[2] In the present study, the TAT 1 and TAT 2 were shortened, which is consistent with previous reports.[5][16] Because the passing autoverification are released immediately to HIS, the TAT is significantly reduced compared with those processed by manual verification, which in turn could save time and decrease the manual workload in clinical laboratories.[6] Furthermore, another benefit is that the autoverification system follows consistent standardized processes, therefore minimizing the potential mistakes made by laboratory technologists due to lack of clinical experience or feelings of fatigue and stress.
This study has limitations to be considered. First, it is difficult to assess the error rate of the autoverification system. Because no obvious mistakes or errors have been reported by patients or clinicians at our hospital for nearly two years, we may infer that the autoverification protocols are at least as safe as the previous manual verification. Second, a higher passing rate of the autoverification system should be designed with future research.
Conclusions
We developed and assessed an autoverification system for the most commonly used coagulation profile with a set of rules. The autoverification system for coagulation assays based on LIS can halt the samples with abnormal values for manual verification, guarantee medical safety, minimize the requirements for manual work, shorten TAT, and raise working efficiency. Furthermore, we desire to design more precise and valuable rules to build a model in which autoverification may best be used on coagulation profiles.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
APTT: Activated partial thromboplastin time
AUTO 10-A: Autoverification of Clinical Laboratory Test Results by CLSI
FBG: Fibrinogen
HIS: Hospital information system
INR: International normalized ratio
LIS: Laboratory information system
PT: Prothrombin time
TAT: Turnaround time
TT: Thrombin time
Acknowledgements
Funding
This research was supported by the Humanities Social Science Foundation of Ministry of Education (Program Grant 17YJCZH184) and Liaoning Education Science 13th Five-Year plan (Program Grant JG17DB557); Philosophy and Social Sciences promotion project of China Medical University (Program Grant 111–3110118083); and The National Natural Science Foundation (Program Grant 81700814). The funding bodies were not involved in neither the design of the study nor the collection, analysis, and interpretation of data, nor the writing of the manuscript.
Author contributions
ZW, HK, and BQ designed the study. ZW, HK, XF, RM, LZ, and MH undertook data collection. CP and BQ analyzed the data. ZW and CP wrote drafts of the manuscript. Then, BQ revised drafts of the paper. All authors contributed to the interpretation of findings and have read and approved the final manuscript.
Ethics declarations
Ethics approval was obtained from the Ethics Committee of the First Affiliated Hospital of China Medical University (2015[23]). The patient consent was waived, as utilization of anonymized history data does not require patient consent.
Competing interests
The authors declare that they have no competing interests.
References
- ↑ Panteghini, M. (2004). "The future of laboratory medicine: understanding the new pressures". The Clinical Biochemist Reviews 25 (4): 207–15. PMC PMC1934959. PMID 18458714. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1934959.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Torke, N.; Boral, L.; Nguyen, T. et al. (2005). "Process improvement and operational efficiency through test result autoverification". Clinical Chemistry 51 (12): 2406–8. doi:10.1373/clinchem.2005.054395. PMID 16306113.
- ↑ 3.0 3.1 3.2 Wu, J.; Pan, M.; Ouyang, H. et al. (2018). "Establishing and Evaluating Autoverification Rules with Intelligent Guidelines for Arterial Blood Gas Analysis in a Clinical Laboratory". SLAS Technology 23 (6): 631–40. doi:10.1177/2472630318775311. PMID 29787327.
- ↑ 4.0 4.1 4.2 4.3 Li, J.; Cheng, B.; Ouyang, H. et al. (2018). "Designing and evaluating autoverification rules for thyroid function profiles and sex hormone tests". Annals of Clinical Biochemistry 55 (2): 254-263. doi:10.1177/0004563217712291. PMID 28490181.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Sediq, A.M.; Abdel-Azeez, A.G. (2014). "Designing an autoverification system in Zagazig University Hospitals Laboratories: Preliminary evaluation on thyroid function profile". Annals of Saudi Medicine 34 (5): 427–32. doi:10.5144/0256-4947.2014.427. PMC PMC6074554. PMID 25827700. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6074554.
- ↑ 6.0 6.1 6.2 6.3 6.4 Randell, E.W.; Short, G.; Lee, N. et al. (2018). "Strategy for 90% autoverification of clinical chemistry and immunoassay test results using six sigma process improvement". Data in Brief 18: 1740-1749. doi:10.1016/j.dib.2018.04.080. PMC PMC5998219. PMID 29904674. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5998219.
- ↑ Jones, J.B. (2013). "A strategic informatics approach to autoverification". Clinics in Laboratory Medicine 33 (1): 161–81. doi:10.1016/j.cll.2012.11.004. PMID 23331736.
- ↑ 8.0 8.1 8.2 Randell, E.W.; Short, G.; Lee, N. et al. (2018). "Autoverification process improvement by Six Sigma approach: Clinical chemistry & immunoassay". Clinical Biochemistry 55: 42–48. doi:10.1016/j.clinbiochem.2018.03.002. PMID 29518383.
- ↑ Gómez-Rioja, R.; Alvarez, V.; Ventura, M. et al. (2013). "Current status of verification practices in clinical biochemistry in Spain". Clinical Chemistry and Laboratory Medicine 51: 1739-46. doi:10.1515/cclm-2012-0659. PMID 23612663.
- ↑ Li, J.; Cheng, B.; Yang, L. et al. (2016). "Development and Implementation of Autoverification Rules for ELISA Results of HBV Serological Markers". Journal of Laboratory Automation 21: 642-51. doi:10.1177/2211068215601612. PMID 26311059.
- ↑ Marsden, N.J.; Van, M.; Dean, S. et al. (2017). "Measuring coagulation in burns: An evidence-based systematic review". Scars, Burns & Healing 3: 2059513117728201. doi:10.1177/2059513117728201. PMC PMC5965330. PMID 29799542. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5965330.
- ↑ Barr, D.; Epps, Q.J. (2019). "Direct oral anticoagulants: A review of common medication errors". Journal of Thrombosis and Thrombolysis 47 (1): 146-154. doi:10.1007/s11239-018-1752-9. PMID 30298305.
- ↑ Favaloro, E.J. (2017). "Optimizing the Verification of Mean Normal Prothrombin Time (MNPT) and International Sensitivity Index (ISI) for Accurate Conversion of Prothrombin Time (PT) to International Normalized Ratio (INR)". Methods in Molecular Biology 1646: 59–74. doi:10.1007/978-1-4939-7196-1_4. PMID 28804818.
- ↑ 14.0 14.1 Gutiérrez García, I.; Pérez Cañadas, P.; Martínez Uriarte, J. et al. (2018). "D-dimer during pregnancy: establishing trimester-specific reference intervals". Scandinavian Journal of Clinical and Laboratory Investigation 78 (6): 439–42. doi:10.1080/00365513.2018.1488177. PMID 29975107.
- ↑ Papageorgiou, C.; Jourdi, G.; Adjambri, E. et al. (2018). "Disseminated Intravascular Coagulation: An Update on Pathogenesis, Diagnosis, and Therapeutic Strategies". Clinical and Applied Thromosis/Hemostasis 24 (9): 8S–28S. doi:10.1177/1076029618806424. PMC PMC6710154. PMID 30296833. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6710154.
- ↑ 16.0 16.1 16.2 16.3 Zhao, Y.; Yang, L.; Zheng, G. et al. (2014). "Building and evaluating the autoverification of coagulation items in the laboratory information system". Clinical Laboratory 60 (1): 143–50. doi:10.1177/1076029618806424. PMID 24600989.
- ↑ 17.0 17.1 "Autoverification of Clinical Laboratory Test Results, 1st Edition". AUTO10. Clinical Laboratory Standards Institute. 31 October 2006. https://clsi.org/standards/products/automation-and-informatics/documents/auto10/.
- ↑ Westgard, J.O. (2003). "Internal quality control: Planning and implementation strategies". Annals of Clinical Biochemistry 40 (Pt 6): 593–611. doi:10.1258/000456303770367199. PMID 14629798.
- ↑ Ovens, K.; Naugler, C. (2012). "How useful are delta checks in the 21 century? A stochastic-dynamic model of specimen mix-up and detection". Journal of Pathology Informatics 3: 5. doi:10.4103/2153-3539.93402. PMC PMC3307229. PMID 22439125. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3307229.
- ↑ 20.0 20.1 Randell, E.W.; Yenice, S. (2019). "Delta Checks in the clinical laboratory". Critical Reviews in Clinical Laboratory Sciences 56 (2): 75-97. doi:10.1080/10408363.2018.1540536. PMID 30632840.
- ↑ Garner, A.E.; Lewington, A.J.; Barth, J.H. (2012). "Detection of patients with acute kidney injury by the clinical laboratory using rises in serum creatinine: Comparison of proposed definitions and a laboratory delta check". Annals of Clinical Biochemistry 49 (Pt 1): 59–62. doi:10.1258/acb.2011.011125. PMID 22130632.
- ↑ 22.0 22.1 Park, S.H.; Kim, S.Y.; Lee, W. et al. (2012). "New decision criteria for selecting delta check methods based on the ratio of the delta difference to the width of the reference range can be generally applicable for each clinical chemistry test item". Annals of Laboratory Medicine 32 (5): 345–54. doi:10.3343/alm.2012.32.5.345. PMC PMC3427822. PMID 22950070. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3427822.
- ↑ 23.0 23.1 Strathmann, F.G.; Baird, G.S.; Hoffman, N.G. (2011). "Simulations of delta check rule performance to detect specimen mislabeling using historical laboratory data". Clinical Chimica Acta 412 (21–22): 1973-7. doi:10.1016/j.cca.2011.07.007. PMID 21782806.
- ↑ Yamashita, T.; Ichihara, K.; Miyamoto, A. (2013). "A novel weighted cumulative delta-check method for highly sensitive detection of specimen mix-up in the clinical laboratory". Clinical Chemistry and Laboratory Medicine 51 (4): 781–9. doi:10.1515/cclm-2012-0752. PMID 23388451.
- ↑ Tran, D.V.; Cembrowski, G.S.; Lee, T. et al. (2008). "Application of 3-D Delta check graphs to HbA1c quality control and HbA1c utilization". American Journal of Clinical Pathology 130 (2): 292-8. doi:10.1309/VM6FVF6GGCYYJ9BV. PMID 18628100.
- ↑ Lacher, D.A.; Connelly, D.P. (1988). "Rate and delta checks compared for selected chemistry tests". Clinical Chemistry 34 (10): 1966–70. PMID 3168205.
- ↑ 27.0 27.1 Xia, L.Y.; Cheng, X.Q.; Liu, Q. et al. (2017). "Developing and application of an autoverification system for clinical chemistry and immunology test results". National Medical Journal of China 97 (8): 616-621. doi:10.3760/cma.j.issn.0376-2491.2017.08.012. PMID 28260308.
- ↑ 28.0 28.1 Krasowski, M.D.; Davis, S.R.; Drees, D. et al. (2014). "Autoverification in a core clinical chemistry laboratory at an academic medical center". Journal of Pathology Informatics 5 (1): 13. doi:10.4103/2153-3539.129450. PMC PMC4023033. PMID 24843824. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4023033.
- ↑ 29.0 29.1 Palmieri, R.; Falbo, R.; Cappellini, F. et al. (2018). "The development of autoverification rules applied to urinalysis performed on the AutionMAX-SediMAX platform". Clinical Chimica Acta 485: 275–81. doi:10.1016/j.cca.2018.07.001. PMID 29981288.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation and grammar, for clarity. In some cases important information was missing from the references, and that information was added.