Difference between revisions of "File:Fig1 OConnor BMCInformatics2010 11-12.jpg"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Added summary.) |
||
| Line 1: | Line 1: | ||
==Summary== | |||
{{Information | |||
|Description='''Figure 1. SeqWare Query Engine schema.''' The HBase database is a generic key-value, column oriented database that pairs well with the inherent sparse matrix nature of variant annotations. (a) The primary table stores multiple genomes worth of generic features, variants, coverages, and variant consequences using genomic location within a particular reference genome as the key. Each genome is represented by a particular column family label (such as “variant:genome7”). For locations with more than one called variant the HBase timestamp is used to distinguish each. (b) Secondary indexing is accomplished using a secondary table per genome indexed. The key is the tag being indexed plus the ID of the object of interest, the value is the row key for the original table. This makes lookup by secondary indexes, “tags” for example, possible without having to iterate over all contents of the primary table. | |||
|Source={{cite journal |url=http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-S12-S2 |title=SeqWare Query Engine: Storing and searching sequence data in the cloud |journal=BMC Informatics |author=O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. |volume=11 |issue=12 |pages=S2 |year=2010 |doi=10.1186/1471-2105-11-S12-S2 |issn=1471-2105}} | |||
|Author=O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. | |||
|Date=2010 | |||
|Permission=[http://creativecommons.org/licenses/by/2.0 Creative Commons Attribution 2.0 Generic] | |||
}} | |||
== Licensing == | == Licensing == | ||
{{cc-by-2.0}} | {{cc-by-2.0}} | ||
Latest revision as of 18:53, 28 December 2015
Summary
| Description |
Figure 1. SeqWare Query Engine schema. The HBase database is a generic key-value, column oriented database that pairs well with the inherent sparse matrix nature of variant annotations. (a) The primary table stores multiple genomes worth of generic features, variants, coverages, and variant consequences using genomic location within a particular reference genome as the key. Each genome is represented by a particular column family label (such as “variant:genome7”). For locations with more than one called variant the HBase timestamp is used to distinguish each. (b) Secondary indexing is accomplished using a secondary table per genome indexed. The key is the tag being indexed plus the ID of the object of interest, the value is the row key for the original table. This makes lookup by secondary indexes, “tags” for example, possible without having to iterate over all contents of the primary table. |
|---|---|
| Source |
O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. (2010). "SeqWare Query Engine: Storing and searching sequence data in the cloud". BMC Informatics 11 (12): S2. doi:10.1186/1471-2105-11-S12-S2. ISSN 1471-2105. http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-S12-S2. |
| Date |
2010 |
| Author |
O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 2.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 18:49, 28 December 2015 | 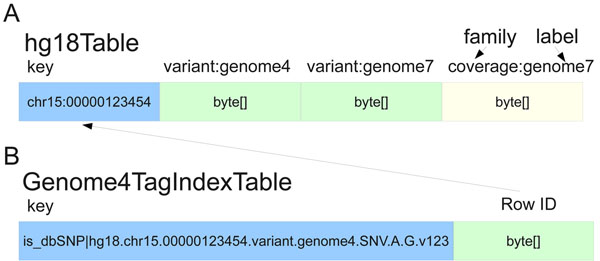 | 600 × 261 (49 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following 2 pages use this file:









