Difference between revisions of "File:Fig1 Stocker BMCBioinformatics2009 10.jpg"
Shawndouglas (talk | contribs) (Added summary.) |
Shawndouglas (talk | contribs) m (→Summary: Spacing) |
||
| Line 2: | Line 2: | ||
{{Information | {{Information | ||
|Description=Figure 1: '''Mapping of the laboratory workflow onto iLAP features.''' The software design of iLAP is inspired by a typical laboratory workflow in life sciences and offers software assistance during the process. The figure illustrates on the left panel the scientific workflow separated into four phases: project definition, data acquisition and analysis, and data retrieval. The right panel shows the main functionalities offered by iLAP. | |Description=Figure 1: '''Mapping of the laboratory workflow onto iLAP features.''' The software design of iLAP is inspired by a typical laboratory workflow in life sciences and offers software assistance during the process. The figure illustrates on the left panel the scientific workflow separated into four phases: project definition, data acquisition and analysis, and data retrieval. The right panel shows the main functionalities offered by iLAP. | ||
|Source={{cite journal |url=http://www.biomedcentral.com/1471-2105/10/390 |title=iLAP: a workflow-driven software for experimental protocol development, data acquisition and analysis |journal=BMC Bioinformatics |author=Stocker, Gernot; Fischer, Maria; Rieder, Dietmar; Bindea, Gabriela; Kainz, Simon; Oberstolz, Michael; McNally, James G.; Trajanoski, Zlatko |volume=10 |pages=390 |year=2009 |doi= | |Source={{cite journal |url=http://www.biomedcentral.com/1471-2105/10/390 |title=iLAP: a workflow-driven software for experimental protocol development, data acquisition and analysis |journal=BMC Bioinformatics |author=Stocker, Gernot; Fischer, Maria; Rieder, Dietmar; Bindea, Gabriela; Kainz, Simon; Oberstolz, Michael; McNally, James G.; Trajanoski, Zlatko |volume=10 |pages=390 |year=2009 |doi=10.1186/1471-2105-10-390 |issn=1471-2105}} | ||
|Author=Stocker, Gernot; Fischer, Maria; Rieder, Dietmar; Bindea, Gabriela; Kainz, Simon; Oberstolz, Michael; McNally, James G.; Trajanoski, Zlatko | |Author=Stocker, Gernot; Fischer, Maria; Rieder, Dietmar; Bindea, Gabriela; Kainz, Simon; Oberstolz, Michael; McNally, James G.; Trajanoski, Zlatko | ||
|Date=2009 | |Date=2009 | ||
Latest revision as of 18:10, 21 January 2016
Summary
| Description |
Figure 1: Mapping of the laboratory workflow onto iLAP features. The software design of iLAP is inspired by a typical laboratory workflow in life sciences and offers software assistance during the process. The figure illustrates on the left panel the scientific workflow separated into four phases: project definition, data acquisition and analysis, and data retrieval. The right panel shows the main functionalities offered by iLAP. |
|---|---|
| Source |
Stocker, Gernot; Fischer, Maria; Rieder, Dietmar; Bindea, Gabriela; Kainz, Simon; Oberstolz, Michael; McNally, James G.; Trajanoski, Zlatko (2009). "iLAP: a workflow-driven software for experimental protocol development, data acquisition and analysis". BMC Bioinformatics 10: 390. doi:10.1186/1471-2105-10-390. ISSN 1471-2105. http://www.biomedcentral.com/1471-2105/10/390. |
| Date |
2009 |
| Author |
Stocker, Gernot; Fischer, Maria; Rieder, Dietmar; Bindea, Gabriela; Kainz, Simon; Oberstolz, Michael; McNally, James G.; Trajanoski, Zlatko |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 2.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 15:42, 19 August 2015 | 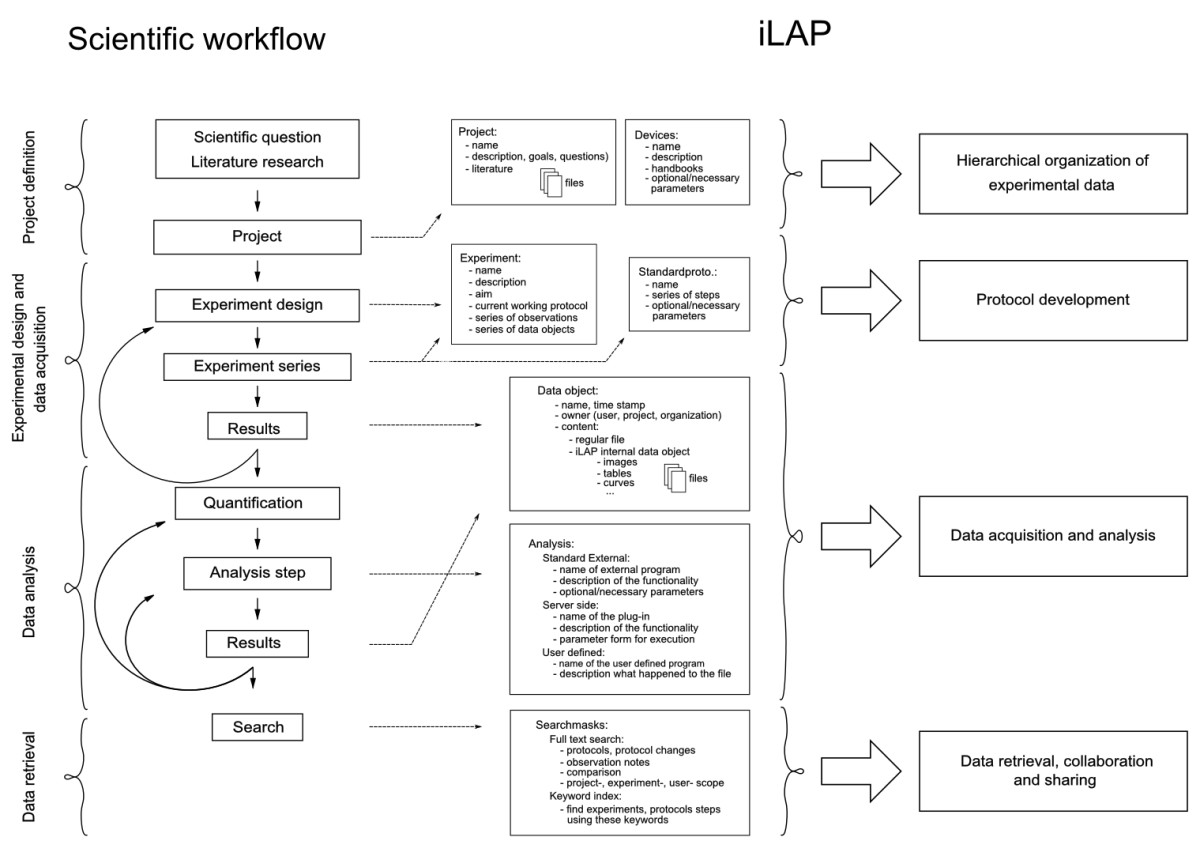 | 1,200 × 856 (117 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









