Difference between revisions of "Journal:Mapping hierarchical file structures to semantic data models for efficient data integration into research data management systems"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Finished adding rest of content) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 18: | Line 18: | ||
|website = [https://www.mdpi.com/2306-5729/9/2/24 https://www.mdpi.com/2306-5729/9/2/24] | |website = [https://www.mdpi.com/2306-5729/9/2/24 https://www.mdpi.com/2306-5729/9/2/24] | ||
|download = [https://www.mdpi.com/2306-5729/9/2/24/pdf?version=1706530315 https://www.mdpi.com/2306-5729/9/2/24/pdf] (PDF) | |download = [https://www.mdpi.com/2306-5729/9/2/24/pdf?version=1706530315 https://www.mdpi.com/2306-5729/9/2/24/pdf] (PDF) | ||
}} | }} | ||
==Abstract== | ==Abstract== | ||
| Line 33: | Line 27: | ||
==Introduction== | ==Introduction== | ||
[[Information management|Data management]] for [[research]] is part of an active transformation in science, with effective management required in order to meet the needs of increasing amounts of complex data. Furthermore, the [[Journal:The FAIR Guiding Principles for scientific data management and stewardship|FAIR guiding principles]] | [[Information management|Data management]] for [[research]] is part of an active transformation in science, with effective management required in order to meet the needs of increasing amounts of complex data. Furthermore, the [[Journal:The FAIR Guiding Principles for scientific data management and stewardship|FAIR guiding principles]]<ref name=":0">{{Cite journal |last=Wilkinson |first=Mark D. |last2=Dumontier |first2=Michel |last3=Aalbersberg |first3=IJsbrand Jan |last4=Appleton |first4=Gabrielle |last5=Axton |first5=Myles |last6=Baak |first6=Arie |last7=Blomberg |first7=Niklas |last8=Boiten |first8=Jan-Willem |last9=da Silva Santos |first9=Luiz Bonino |last10=Bourne |first10=Philip E. |last11=Bouwman |first11=Jildau |date=2016-03-15 |title=The FAIR Guiding Principles for scientific data management and stewardship |url=https://www.nature.com/articles/sdata201618 |journal=Scientific Data |language=en |volume=3 |issue=1 |pages=160018 |doi=10.1038/sdata.2016.18 |issn=2052-4463 |pmc=PMC4792175 |pmid=26978244}}</ref> for scientific data—which are an elementary part of numerous data management plans, funding guidelines, and data management strategies of research organizations<ref>{{Cite journal |last=Deutsche Forschungsgemeinschaft |date=2022-04-20 |title=Guidelines for Safeguarding Good Research Practice. Code of Conduct |url=https://zenodo.org/record/6472827 |journal=Zenodo |language=de |doi=10.5281/ZENODO.6472827}}</ref><ref>{{Cite journal |last=Ferguson |first=Lea Maria |last2=Bertelmann |first2=Roland |last3=Bruch |first3=Christoph |last4=Messerschmidt |first4=Reinhard |last5=Pampel |first5=Heinz |last6=Schrader |first6=Antonia |last7=Schultze-Motel |first7=Paul |last8=Weisweiler |first8=Nina Leonie |date=2022 |title=Good (Digital) Research Practice and Open Science Support and Best Practices for Implementing the DFG Code of Conduct “Guidelines for Safeguarding Good Research Practice”. Helmholtz Open Science Briefing. Good (Digital) Research Practice and Open Science Support and Best Practices for Implementing the DFG Code of Conduct “Guidelines for Safeguarding Good Research Practice” Version 2.0 |url=https://juser.fz-juelich.de/record/1014764 |language=en |pages=pages 19 pages470 kb |doi=10.34734/fzj-2023-03453}}</ref>, requiring that research objects be more findable, accessible, interoperable, and reusable—require scientists to review and enhance their established data management [[workflow]]s. | ||
One particular focus of this endeavor is the introduction and expansion of research data management systems (RDMSs). These systems help researchers organize their data during the whole data management life cycle, especially by increasing findability and accessibility. | One particular focus of this endeavor is the introduction and expansion of research data management systems (RDMSs). These systems help researchers organize their data during the whole data management life cycle, especially by increasing findability and accessibility.<ref name=":1">{{Cite journal |last=Gray |first=Jim |last2=Liu |first2=David T. |last3=Nieto-Santisteban |first3=Maria |last4=Szalay |first4=Alex |last5=DeWitt |first5=David J. |last6=Heber |first6=Gerd |date=2005-12 |title=Scientific data management in the coming decade |url=https://dl.acm.org/doi/10.1145/1107499.1107503 |journal=ACM SIGMOD Record |language=en |volume=34 |issue=4 |pages=34–41 |doi=10.1145/1107499.1107503 |issn=0163-5808}}</ref> Furthermore, [[Semantics|semantic]] data management approaches<ref>{{Citation |last=Samuel |first=Sheeba |date=2017 |editor-last=Blomqvist |editor-first=Eva |editor2-last=Maynard |editor2-first=Diana |editor3-last=Gangemi |editor3-first=Aldo |editor4-last=Hoekstra |editor4-first=Rinke |editor5-last=Hitzler |editor5-first=Pascal |title=Integrative Data Management for Reproducibility of Microscopy Experiments |url=https://link.springer.com/10.1007/978-3-319-58451-5_19 |work=The Semantic Web |language=en |publisher=Springer International Publishing |place=Cham |volume=10250 |pages=246–255 |doi=10.1007/978-3-319-58451-5_19 |isbn=978-3-319-58450-8 |accessdate=2024-06-11}}</ref> can increase the reuse and reproducibility of data that are typically organized in file structures. As has been pointed out by Gray ''et al.''<ref name=":1" />, one major shortcoming of file systems is the lack of rich [[metadata]] features, which additionally limits search options. Typically, RDMSs employ [[database]] management systems (DBMSs) to store data and metadata, but the degree to which data is migrated, linked, or synchronized into these systems can vary substantially. | ||
The import of data into an RDMS typically requires the development of [[data integration]] procedures that are tied to the specific workflows at hand. While very few standard products exist | The import of data into an RDMS typically requires the development of [[data integration]] procedures that are tied to the specific workflows at hand. While very few standard products exist<ref name=":2">{{Cite book |last=Vaisman |first=Alejandro |last2=Zimányi |first2=Esteban |date=2014 |title=Data Warehouse Systems: Design and Implementation |url=https://link.springer.com/10.1007/978-3-642-54655-6 |language=en |publisher=Springer Berlin Heidelberg |place=Berlin, Heidelberg |doi=10.1007/978-3-642-54655-6 |isbn=978-3-642-54654-9}}</ref>, in practice, mostly custom software written in various programming languages and making use of a high variety of different software packages are used for data integration in scientific environments. There are two main workflows for integrating data into RDMSs: manually inputting data (e.g., using forms<ref>{{Cite journal |last=Barillari |first=Caterina |last2=Ottoz |first2=Diana S. M. |last3=Fuentes-Serna |first3=Juan Mariano |last4=Ramakrishnan |first4=Chandrasekhar |last5=Rinn |first5=Bernd |last6=Rudolf |first6=Fabian |date=2016-02-15 |title=openBIS ELN-LIMS: an open-source database for academic laboratories |url=https://academic.oup.com/bioinformatics/article/32/4/638/1743839 |journal=Bioinformatics |language=en |volume=32 |issue=4 |pages=638–640 |doi=10.1093/bioinformatics/btv606 |issn=1367-4811 |pmc=PMC4743625 |pmid=26508761}}</ref>) or facilitating the batch import of data sets. The automatic methods often include data import routines for predefined formats, like tables in Excel or .csv format.<ref>{{Cite journal |last=Hewera |first=Michael |last2=Hänggi |first2=Daniel |last3=Gerlach |first3=Björn |last4=Kahlert |first4=Ulf Dietrich |date=2021-08-02 |title=eLabFTW as an Open Science tool to improve the quality and translation of preclinical research |url=https://f1000research.com/articles/10-292/v3 |journal=F1000Research |language=en |volume=10 |pages=292 |doi=10.12688/f1000research.52157.3 |issn=2046-1402 |pmc=PMC8323070 |pmid=34381592}}</ref><ref>{{Cite journal |last=Suhr |first=M. |last2=Lehmann |first2=C. |last3=Bauer |first3=C. R. |last4=Bender |first4=T. |last5=Knopp |first5=C. |last6=Freckmann |first6=L. |last7=Öst Hansen |first7=B. |last8=Henke |first8=C. |last9=Aschenbrandt |first9=G. |last10=Kühlborn |first10=L. K. |last11=Rheinländer |first11=S. |date=2020-12 |title=Menoci: lightweight extensible web portal enhancing data management for biomedical research projects |url=https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03928-1 |journal=BMC Bioinformatics |language=en |volume=21 |issue=1 |pages=582 |doi=10.1186/s12859-020-03928-1 |issn=1471-2105 |pmc=PMC7745495 |pmid=33334310}}</ref> Some systems include plugin systems to allow for a configuration of the data integration process.<ref>{{Cite journal |last=Bauch |first=Angela |last2=Adamczyk |first2=Izabela |last3=Buczek |first3=Piotr |last4=Elmer |first4=Franz-Josef |last5=Enimanev |first5=Kaloyan |last6=Glyzewski |first6=Pawel |last7=Kohler |first7=Manuel |last8=Pylak |first8=Tomasz |last9=Quandt |first9=Andreas |last10=Ramakrishnan |first10=Chandrasekhar |last11=Beisel |first11=Christian |date=2011-12 |title=openBIS: a flexible framework for managing and analyzing complex data in biology research |url=https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-468 |journal=BMC Bioinformatics |language=en |volume=12 |issue=1 |pages=468 |doi=10.1186/1471-2105-12-468 |issn=1471-2105 |pmc=PMC3275639 |pmid=22151573}}</ref> Sometimes, data files have to be uploaded using a web front-end<ref>{{Citation |last=Dudchenko |first=Aleksei |last2=Ringwald |first2=Friedemann |last3=Czernilofsky |first3=Felix |last4=Dietrich |first4=Sascha |last5=Knaup |first5=Petra |last6=Ganzinger |first6=Matthias |date=2022-05-25 |editor-last=Séroussi |editor-first=Brigitte |editor2-last=Weber |editor2-first=Patrick |editor3-last=Dhombres |editor3-first=Ferdinand |editor4-last=Grouin |editor4-first=Cyril |editor5-last=Liebe |editor5-first=Jan-David |title=Large-File Raw Data Synchronization for openBIS Research Repositories |url=https://ebooks.iospress.nl/doi/10.3233/SHTI220486 |work=Studies in Health Technology and Informatics |publisher=IOS Press |doi=10.3233/shti220486 |isbn=978-1-64368-284-6 |accessdate=2024-06-11}}</ref> and are afterwards attached to objects in the RDMSs. In general, developing this kind of software can be considered very costly<ref name=":2" />, as it is highly dependent on the specific environment. Data import can still be considered one of the major bottlenecks for the adaption of an RDMS. | ||
There are several advantages to using an RDMS over organization of data in classical file hierarchies. There is a higher flexibility in adding metadata to data sets, while these capabilities are limited for classical file systems. The standardized representation in an RDMS improves the comparability of data sets that possibly originate from different file formats and data representations. Furthermore, semantic information can be seamlessly integrated, possibly using standards like RDF | There are several advantages to using an RDMS over organization of data in classical file hierarchies. There is a higher flexibility in adding metadata to data sets, while these capabilities are limited for classical file systems. The standardized representation in an RDMS improves the comparability of data sets that possibly originate from different file formats and data representations. Furthermore, semantic information can be seamlessly integrated, possibly using standards like RDF<ref>{{Citation |last=McBride |first=Brian |date=2004 |editor-last=Staab |editor-first=Steffen |editor2-last=Studer |editor2-first=Rudi |title=The Resource Description Framework (RDF) and its Vocabulary Description Language RDFS |url=http://link.springer.com/10.1007/978-3-540-24750-0_3 |work=Handbook on Ontologies |language=en |publisher=Springer Berlin Heidelberg |place=Berlin, Heidelberg |pages=51–65 |doi=10.1007/978-3-540-24750-0_3 |isbn=978-3-662-11957-0 |accessdate=2024-06-11}}</ref> and OWL.<ref>{{Cite web |last=W3C OWL Working Group |date=11 December 2012 |title=OWL 2 Web Ontology Language Document Overview (Second Edition) |url=https://www.w3.org/TR/owl2-overview/ |publisher=World Wide Web Consortium}}</ref> The semantic information allows for advanced querying and searching, e.g., using SPARQL.<ref>{{Cite journal |last=Pérez |first=Jorge |last2=Arenas |first2=Marcelo |last3=Gutierrez |first3=Claudio |date=2009-08 |title=Semantics and complexity of SPARQL |url=https://dl.acm.org/doi/10.1145/1567274.1567278 |journal=ACM Transactions on Database Systems |language=en |volume=34 |issue=3 |pages=1–45 |doi=10.1145/1567274.1567278 |issn=0362-5915}}</ref> Concepts like linked data<ref>{{Cite web |last=Bizer, C.; Heath, T.; Ayers, D. et al. |date=2007 |title=Interlinking Open Data on the Web |work=Proceedings of the 4th European Semantic Web Conference |url=http://tomheath.com/papers/bizer-heath-eswc2007-interlinking-open-data.pdf |format=PDF}}</ref><ref>{{Cite journal |last=Bizer |first=Christian |last2=Heath |first2=Tom |last3=Berners-Lee |first3=Tim |date=2009-07-01 |title=Linked Data - The Story So Far: |url=https://services.igi-global.com/resolvedoi/resolve.aspx?doi=10.4018/jswis.2009081901 |journal=International Journal on Semantic Web and Information Systems |language=ng |volume=5 |issue=3 |pages=1–22 |doi=10.4018/jswis.2009081901 |issn=1552-6283}}</ref> and FAIR digital objects (FDO<ref name=":3">{{Cite journal |last=De Smedt |first=Koenraad |last2=Koureas |first2=Dimitris |last3=Wittenburg |first3=Peter |date=2020-04-11 |title=FAIR Digital Objects for Science: From Data Pieces to Actionable Knowledge Units |url=https://www.mdpi.com/2304-6775/8/2/21 |journal=Publications |language=en |volume=8 |issue=2 |pages=21 |doi=10.3390/publications8020021 |issn=2304-6775}}</ref>) provide overarching concepts for achieving more standardized representations within RDMSs and for publication on the web. Specifically, the FDO concept aims at bundling data sets with a persistent identifier (PID) and its metadata to self-contained units. These units are designed to be machine-actionable and interoperable, so that they have the potential to build complex and distributed data processing infrastructures.<ref name=":3" /> | ||
===Using file systems and RDMSs simultaneously=== | ===Using file systems and RDMSs simultaneously=== | ||
Despite the advantages mentioned above, RDMSs have still failed to gain a widespread adoption. One of the key problems in the employment of an RDMS in an active research environment is that a full transition to such a system is very difficult, as most digital scientific workflows are in one or multiple ways dependent on classical hierarchical file systems. | Despite the advantages mentioned above, RDMSs have still failed to gain a widespread adoption. One of the key problems in the employment of an RDMS in an active research environment is that a full transition to such a system is very difficult, as most digital scientific workflows are in one or multiple ways dependent on classical hierarchical file systems.<ref name=":1" /> Examples include data acquisition and measurement devices, data processing and analysis software, and digitized [[laboratory]] notes and material for publications. The complete transition to an RDMS would require developing data integration procedures (e.g., extract, transform, load [ETL]<ref name=":2" /><ref>{{Cite journal |last=Vassiliadis |first=Panos |date=2009-07-01 |title=A Survey of Extract–Transform–Load Technology: |url=https://services.igi-global.com/resolvedoi/resolve.aspx?doi=10.4018/jdwm.2009070101 |journal=International Journal of Data Warehousing and Mining |language=ng |volume=5 |issue=3 |pages=1–27 |doi=10.4018/jdwm.2009070101 |issn=1548-3924}}</ref> processes) for every digital workflow in the lab and to provide interfaces for input and output to any other software involved in these workflows. | ||
As files on classical file systems play a crucial role in these workflows, our aim is to develop a robust strategy to use file systems and an RDMS simultaneously. Rather than requiring a full transition to an RDMS, we want to make use of the file system as an interoperability layer between the RDMS and any other file-based workflow in the research environment. | As files on classical file systems play a crucial role in these workflows, our aim is to develop a robust strategy to use file systems and an RDMS simultaneously. Rather than requiring a full transition to an RDMS, we want to make use of the file system as an interoperability layer between the RDMS and any other file-based workflow in the research environment. | ||
| Line 53: | Line 47: | ||
Apart from the main motivation, described above, we have identified several additional advantages of using a conventional folder structure simultaneous to an RDMS: standard tools for managing the files can be used for [[backup]] (e.g., rsync), [[Version control|versioning]] (e.g., git), archiving, and file access (e.g., SSH). Functionality of these tools does not need to be re-implemented in the RDMS. Furthermore, the file system can act as a fallback in cases where the RDMS might become unavailable. This methodology, therefore, increases robustness. As a third advantage, existing workflows relying on storing files in a file system do not need to be changed, while the simultaneous findability within an RDMS is available to users. | Apart from the main motivation, described above, we have identified several additional advantages of using a conventional folder structure simultaneous to an RDMS: standard tools for managing the files can be used for [[backup]] (e.g., rsync), [[Version control|versioning]] (e.g., git), archiving, and file access (e.g., SSH). Functionality of these tools does not need to be re-implemented in the RDMS. Furthermore, the file system can act as a fallback in cases where the RDMS might become unavailable. This methodology, therefore, increases robustness. As a third advantage, existing workflows relying on storing files in a file system do not need to be changed, while the simultaneous findability within an RDMS is available to users. | ||
The concepts described in this article can be used independent of a specific RDMS software. However, as a proof-of-concept, we implemented the approach as part of the file crawler framework that belongs to the open-source RDMS LinkAhead (recently renamed from CaosDB). | The concepts described in this article can be used independent of a specific RDMS software. However, as a proof-of-concept, we implemented the approach as part of the file crawler framework that belongs to the open-source RDMS LinkAhead (recently renamed from CaosDB).<ref name=":4">{{Cite journal |last=Fitschen |first=Timm |last2=Schlemmer |first2=Alexander |last3=Hornung |first3=Daniel |last4=tom Wörden |first4=Henrik |last5=Parlitz |first5=Ulrich |last6=Luther |first6=Stefan |date=2019-06-10 |title=CaosDB—Research Data Management for Complex, Changing, and Automated Research Workflows |url=https://www.mdpi.com/2306-5729/4/2/83 |journal=Data |language=en |volume=4 |issue=2 |pages=83 |doi=10.3390/data4020083 |issn=2306-5729}}</ref><ref>{{Cite journal |last=Hornung |first=Daniel |last2=Spreckelsen |first2=Florian |last3=Weiß |first3=Thomas |date=2024-01-30 |title=Agile Research Data Management with Open Source: LinkAhead |url=https://www.inggrid.org/article/id/3866/ |language=en |doi=10.48694/INGGRID.3866 |issn=2941-1300}}</ref> The crawler framework is released as [[open-source software]] under the AGPLv3 license (see Appendix A). | ||
===Example data set=== | ===Example data set=== | ||
We will illustrate the problem of integrating research data using a simplified example that is based on the work of Spreckelsen ''et al.'' | We will illustrate the problem of integrating research data using a simplified example that is based on the work of Spreckelsen ''et al.''<ref name=":5">{{Cite journal |last=Spreckelsen |first=Florian |last2=Rüchardt |first2=Baltasar |last3=Lebert |first3=Jan |last4=Luther |first4=Stefan |last5=Parlitz |first5=Ulrich |last6=Schlemmer |first6=Alexander |date=2020-04-24 |title=Guidelines for a Standardized Filesystem Layout for Scientific Data |url=https://www.mdpi.com/2306-5729/5/2/43 |journal=Data |language=en |volume=5 |issue=2 |pages=43 |doi=10.3390/data5020043 |issn=2306-5729}}</ref> This example will be used in the results section to demonstrate our data integration concepts. Examples for more complex data integration, e.g., for data sets found in the neurosciences (BIDS<ref>{{Cite journal |last=Gorgolewski |first=Krzysztof J. |last2=Auer |first2=Tibor |last3=Calhoun |first3=Vince D. |last4=Craddock |first4=R. Cameron |last5=Das |first5=Samir |last6=Duff |first6=Eugene P. |last7=Flandin |first7=Guillaume |last8=Ghosh |first8=Satrajit S. |last9=Glatard |first9=Tristan |last10=Halchenko |first10=Yaroslav O. |last11=Handwerker |first11=Daniel A. |date=2016-06-21 |title=The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments |url=https://www.nature.com/articles/sdata201644 |journal=Scientific Data |language=en |volume=3 |issue=1 |pages=160044 |doi=10.1038/sdata.2016.44 |issn=2052-4463 |pmc=PMC4978148 |pmid=27326542}}</ref> and DICOM<ref>{{Cite journal |last=Mildenberger |first=Peter |last2=Eichelberg |first2=Marco |last3=Martin |first3=Eric |date=2002-04 |title=Introduction to the DICOM standard |url=http://link.springer.com/10.1007/s003300101100 |journal=European Radiology |language=en |volume=12 |issue=4 |pages=920–927 |doi=10.1007/s003300101100 |issn=0938-7994}}</ref>) and in the geosciences, can be found online (see Appendix B). Although the concept is not restricted to data stored on file systems, in this example we will assume for simplicity that the research data are stored on a standard file system with a well-defined file structure layout: | ||
<tt>ExperimentalData/<br /> | <tt>ExperimentalData/<br /> | ||
:2020_SpeedOfLight/<br /> | :2020_SpeedOfLight/<br /> | ||
::2020-01-01_TimeOfFlight<br /> | ::2020-01-01_TimeOfFlight<br /> | ||
:::README.md<br /> | :::README.md<br /> | ||
| Line 66: | Line 62: | ||
:::README.md<br /> | :::README.md<br /> | ||
:::...<br /> | :::...<br /> | ||
::2020-01-03<br /> | ::2020-01-03<br /> | ||
:::README.md<br /> | :::README.md<br /> | ||
:::...</tt> | :::...</tt> | ||
The above listing replicates an example with experimental data from Spreckelsen ''et al.'' | The above listing replicates an example with experimental data from Spreckelsen ''et al.''<ref name=":5" /> using a three-level folder structure: | ||
* Level 1 (ExperimentalData) stores rough categories for data, in this data acquired from experimental measurements. | *Level 1 (ExperimentalData) stores rough categories for data, in this data acquired from experimental measurements. | ||
* Level 2 (2020_SpeedOfLight) is the level of project names, grouping data into independent projects. | *Level 2 (2020_SpeedOfLight) is the level of project names, grouping data into independent projects. | ||
* Level 3 stores the actual measurement folders, which can also be referred to as “scientific activity” folders in the general case. Each of these folders could have an arbitrary substructure and store the actual experimental data along with a README.md file, containing meta data. | *Level 3 stores the actual measurement folders, which can also be referred to as “scientific activity” folders in the general case. Each of these folders could have an arbitrary substructure and store the actual experimental data along with a README.md file, containing meta data. | ||
The generic use case of integrating data from file systems involves the following sub tasks: | The generic use case of integrating data from file systems involves the following sub tasks: | ||
| Line 90: | Line 88: | ||
====Data models in LinkAhead==== | ====Data models in LinkAhead==== | ||
The LinkAhead data model is basically an object-oriented representation of data which makes use of four different types of entities: <tt>RecordType</tt>, <tt>Property</tt>, <tt>Record</tt> and <tt>File RecordTypes</tt>, and <tt>Properties</tt>. These four entities define the data model, which is later used to store concrete data objects, which are represented by <tt>Records</tt>. In that respect, <tt>RecordTypes</tt> and <tt>Properties</tt> share a lot of similarities with [[Ontology (information science)|ontologies]], but have a restricted set of relations, as described in more detail by Fitschen ''et al.'' | The LinkAhead data model is basically an object-oriented representation of data which makes use of four different types of entities: <tt>RecordType</tt>, <tt>Property</tt>, <tt>Record</tt> and <tt>File RecordTypes</tt>, and <tt>Properties</tt>. These four entities define the data model, which is later used to store concrete data objects, which are represented by <tt>Records</tt>. In that respect, <tt>RecordTypes</tt> and <tt>Properties</tt> share a lot of similarities with [[Ontology (information science)|ontologies]], but have a restricted set of relations, as described in more detail by Fitschen ''et al.''<ref name=":4" /> <tt>Files</tt> have a special role within LinkAhead as they represent references to actual files on a file system, but allow for linking them to other LinkAhead entities and, e.g., adding custom properties. | ||
<tt>Properties</tt> are individual pieces of information that have a name, description, optionally a physical unit, and can store a value of a well-defined data type. <tt>Properties</tt> are attached to <tt>RecordTypes</tt> and can be marked as “obligatory,” “recommended,” or “suggested.” In case of obligatory <tt>Properties</tt>, each <tt>Record</tt> of the respective <tt>RecordType</tt> is enforced to set the respective <tt>Properties</tt>. Each Record must have at least one <tt>RecordType</tt> and <tt>RecordTypes</tt> can have other <tt>RecordTypes</tt> as parents. This is known as (multiple) inheritance in object-oriented programming languages. | <tt>Properties</tt> are individual pieces of information that have a name, description, optionally a physical unit, and can store a value of a well-defined data type. <tt>Properties</tt> are attached to <tt>RecordTypes</tt> and can be marked as “obligatory,” “recommended,” or “suggested.” In case of obligatory <tt>Properties</tt>, each <tt>Record</tt> of the respective <tt>RecordType</tt> is enforced to set the respective <tt>Properties</tt>. Each Record must have at least one <tt>RecordType</tt> and <tt>RecordTypes</tt> can have other <tt>RecordTypes</tt> as parents. This is known as (multiple) inheritance in object-oriented programming languages. | ||
| Line 100: | Line 98: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="700px" | {| border="0" cellpadding="5" cellspacing="0" width="700px" | ||
|- | |- | ||
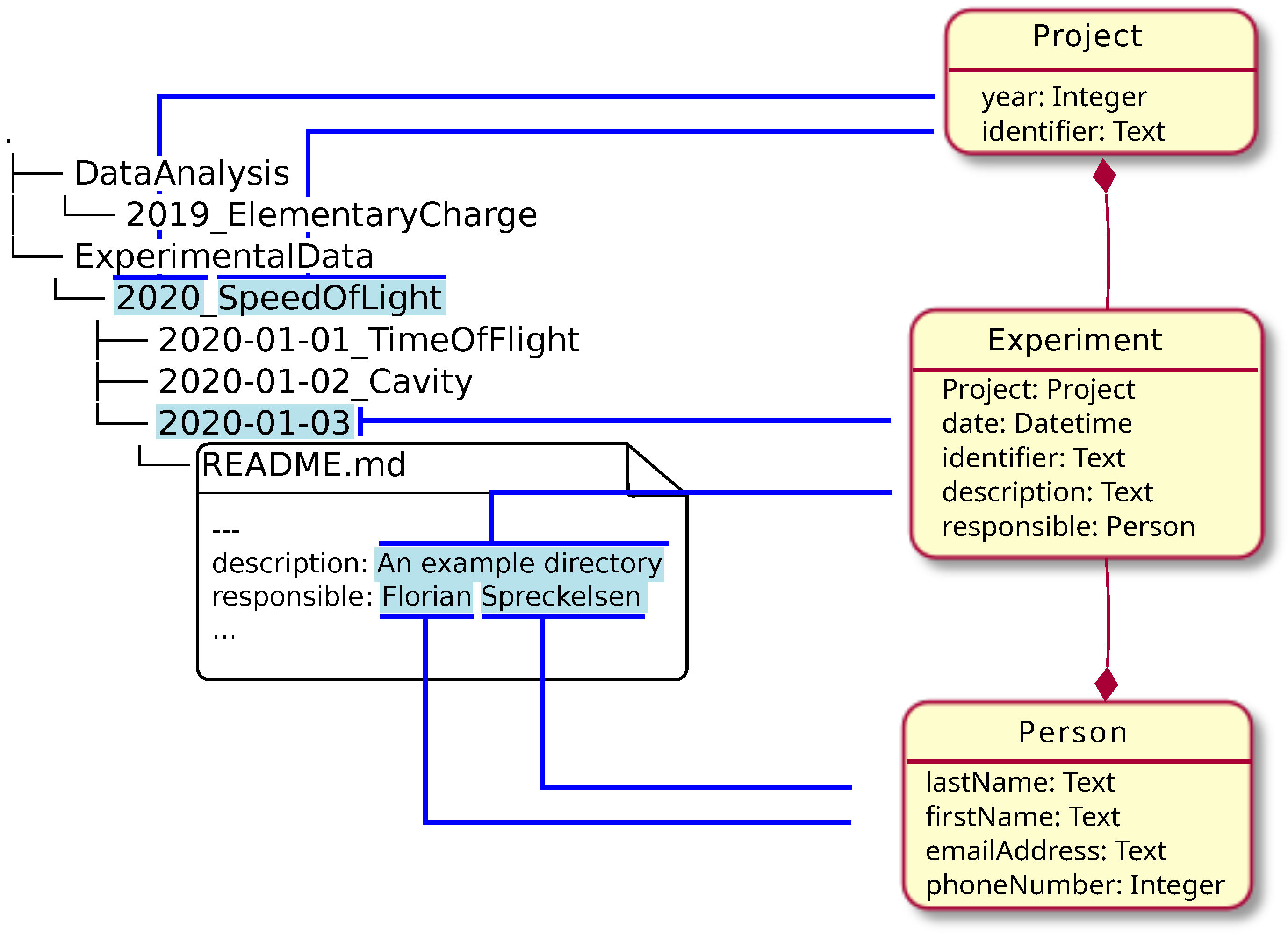
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1.''' Mapping between file structure and data model. Blue lines indicate which pieces of information from the file structure and file contents are mapped to the respective properties in the data model.</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 1.''' Mapping between file structure and data model. Blue lines indicate which pieces of information from the file structure and file contents are mapped to the respective properties in the data model.</blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 120: | Line 118: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="700px" | {| border="0" cellpadding="5" cellspacing="0" width="700px" | ||
|- | |- | ||
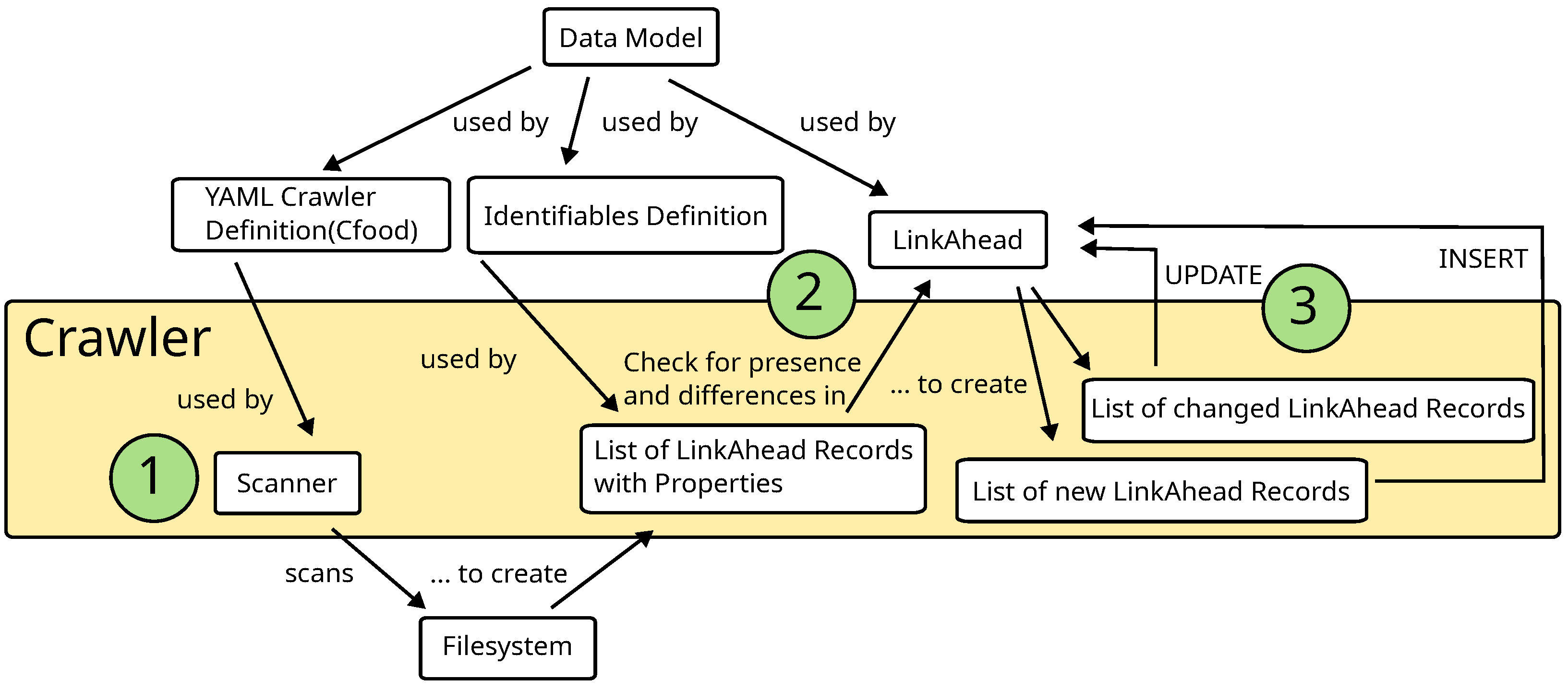
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2.''' Overview of the complete data integration procedure.</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 2.''' Overview of the complete data integration procedure.</blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 129: | Line 127: | ||
===Mapping files and layouts into a data model=== | ===Mapping files and layouts into a data model=== | ||
Suppose we have a hierarchical structure of some kind that contains certain information and we want to map this information onto our object-oriented data model. The typical example for the hierarchical structure would be a folder structure with files, but hierarchical data formats like HDF5 | Suppose we have a hierarchical structure of some kind that contains certain information and we want to map this information onto our object-oriented data model. The typical example for the hierarchical structure would be a folder structure with files, but hierarchical data formats like HDF5<ref name=":6">{{Citation |last=Koranne |first=Sandeep |date=2011 |title=Hierarchical Data Format 5 : HDF5 |url=https://link.springer.com/10.1007/978-1-4419-7719-9_10 |work=Handbook of Open Source Tools |language=en |publisher=Springer US |place=Boston, MA |pages=191–200 |doi=10.1007/978-1-4419-7719-9_10 |isbn=978-1-4419-7718-2 |accessdate=2024-06-11}}</ref><ref name=":7">{{Cite journal |last=Folk |first=Mike |last2=Heber |first2=Gerd |last3=Koziol |first3=Quincey |last4=Pourmal |first4=Elena |last5=Robinson |first5=Dana |date=2011-03-25 |title=An overview of the HDF5 technology suite and its applications |url=https://dl.acm.org/doi/10.1145/1966895.1966900 |journal=Proceedings of the EDBT/ICDT 2011 Workshop on Array Databases |language=en |publisher=ACM |place=Uppsala Sweden |pages=36–47 |doi=10.1145/1966895.1966900 |isbn=978-1-4503-0614-0}}</ref> files (or a mixture of both) would also fit the use case. | ||
The prior Figure 1 illustrates what such a mapping could look like in practice using the prior stated file structure: | The prior Figure 1 illustrates what such a mapping could look like in practice using the prior stated file structure: | ||
* ExperimentalData contains one subfolder 2020_SpeedOfLight, storing all data from this experimental series. The experimental series is represented in a <tt>RecordType</tt> called <tt>Project</tt>. The <tt>Properties</tt> “year” (2020) and “identifier” (SpeedOfLight) can be directly filled from the directory name. | *ExperimentalData contains one subfolder 2020_SpeedOfLight, storing all data from this experimental series. The experimental series is represented in a <tt>RecordType</tt> called <tt>Project</tt>. The <tt>Properties</tt> “year” (2020) and “identifier” (SpeedOfLight) can be directly filled from the directory name. | ||
* Each experiment of the series has its dedicated subfolder, so one <tt>Record</tt> of type <tt>Experiment</tt> will be created for each one. The association to its <tt>Project</tt>, which is implicitly clear in the folder hierarchy, can be mapped to a reference to the <tt>Project Record</tt> created in the previous step. Again, the <tt>Property</tt> “date” can be set from the directory name. | *Each experiment of the series has its dedicated subfolder, so one <tt>Record</tt> of type <tt>Experiment</tt> will be created for each one. The association to its <tt>Project</tt>, which is implicitly clear in the folder hierarchy, can be mapped to a reference to the <tt>Project Record</tt> created in the previous step. Again, the <tt>Property</tt> “date” can be set from the directory name. | ||
* Each experiment contains a file called README.md storing text and metadata about the experiment, according to the described standard. In this case, the “description” <tt>Property</tt> for the <tt>Experiment Record</tt> created in the previous step will be set from the corresponding YAML value. Furthermore, a <tt>Person Record</tt> with <tt>Properties</tt> "firstName = Florian" and "lastName = Spreckelsen" will be created. Finally the <tt>Person Record</tt> will be set as the value for the property “responsible” of the <tt>Experiment Record</tt>. | *Each experiment contains a file called README.md storing text and metadata about the experiment, according to the described standard. In this case, the “description” <tt>Property</tt> for the <tt>Experiment Record</tt> created in the previous step will be set from the corresponding YAML value. Furthermore, a <tt>Person Record</tt> with <tt>Properties</tt> "firstName = Florian" and "lastName = Spreckelsen" will be created. Finally the <tt>Person Record</tt> will be set as the value for the property “responsible” of the <tt>Experiment Record</tt>. | ||
====YAML definitions==== | ====YAML definitions==== | ||
| Line 144: | Line 142: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width=" | {| border="0" cellpadding="5" cellspacing="0" width="800px" | ||
|- | |- | ||
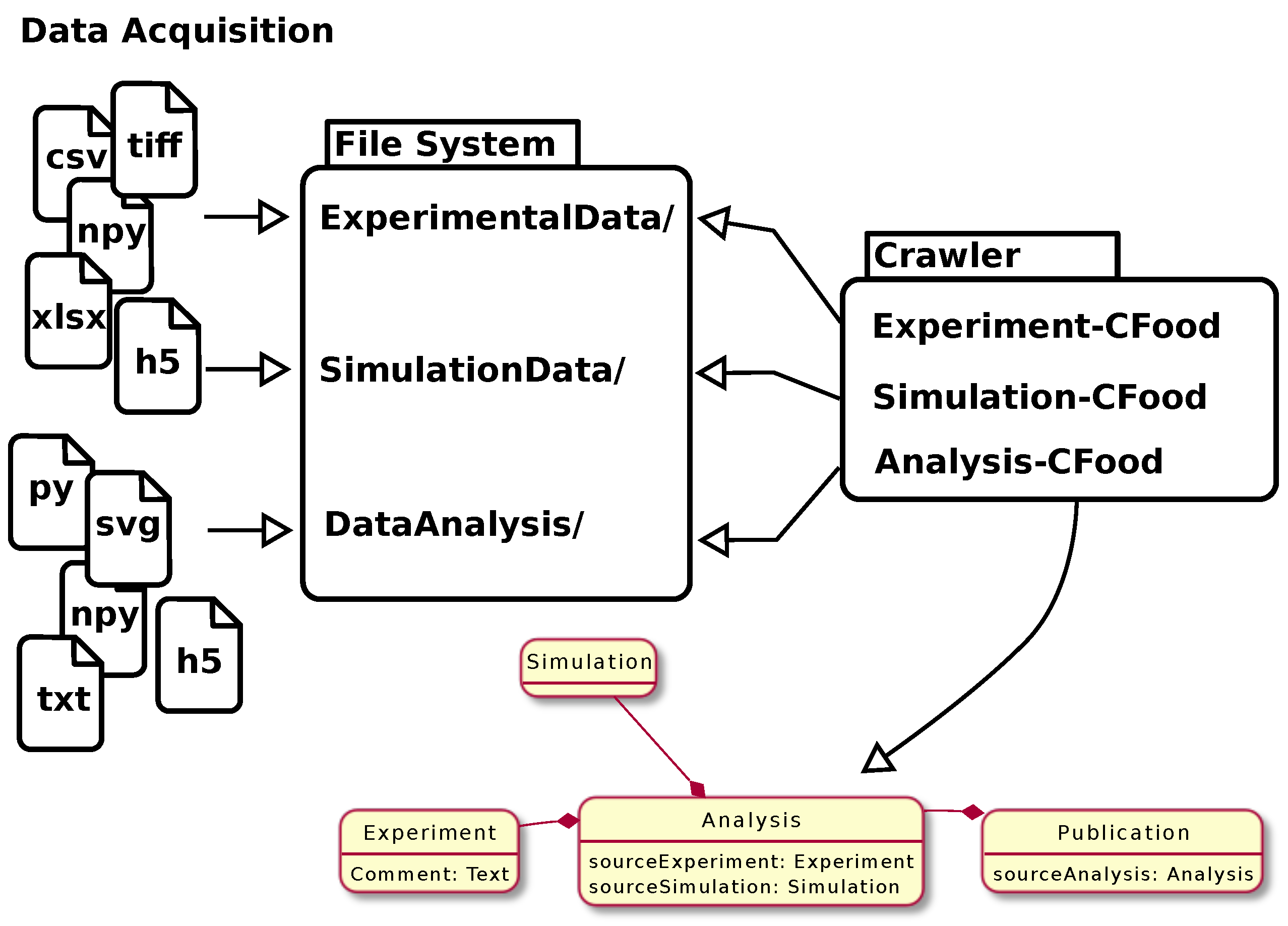
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 3.''' Illustration of the data synchronization procedure using a crawler. Data acquisition (possibly including computer simulations or data produced during data analysis) leads to a variety of files in different formats on the file system. Crawler plugins (<tt>CFoods</tt>) are designed in a way that they understand the local structures on the file system. The file tree is traversed, possibly opening files and extracting (meta) data in order to transform them into a semantic data model that is then synchronized with the RDMS. The figure was previously published by Schlemmer. | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 3.''' Illustration of the data synchronization procedure using a crawler. Data acquisition (possibly including computer simulations or data produced during data analysis) leads to a variety of files in different formats on the file system. Crawler plugins (<tt>CFoods</tt>) are designed in a way that they understand the local structures on the file system. The file tree is traversed, possibly opening files and extracting (meta) data in order to transform them into a semantic data model that is then synchronized with the RDMS. The figure was previously published by Schlemmer.<ref>{{Citation |last=Schlemmer, Alexander |date=2021-05-05 |title=Mapping data files to semantic data models using the CaosDB crawler |url=https://zenodo.org/record/8246645 |work=Zenodo |language=en |doi=10.5281/zenodo.8246645 |accessdate=2024-06-11}}</ref></blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 197: | Line 195: | ||
::::::::::Experiment:<br /> | ::::::::::Experiment:<br /> | ||
:::::::::::Person: $Person | :::::::::::Person: $Person | ||
</tt> | </tt> | ||
| Line 203: | Line 202: | ||
In the given example, the following converters can be found: | In the given example, the following converters can be found: | ||
* ExperimentalData_Dir, Project_Dir, Experiment_Dir, which are all of type <tt>Directory</tt>. These converters match names of folders against the regular expression given by match. When matching, converters of this type yield sub-folders and -files for processing of converters in the subtree section. | *ExperimentalData_Dir, Project_Dir, Experiment_Dir, which are all of type <tt>Directory</tt>. These converters match names of folders against the regular expression given by match. When matching, converters of this type yield sub-folders and -files for processing of converters in the subtree section. | ||
* Readme_File is of type <tt>MarkdownFile</tt> and allows for processing of the contents of the YAML header by converters given in the subtree section. | *Readme_File is of type <tt>MarkdownFile</tt> and allows for processing of the contents of the YAML header by converters given in the subtree section. | ||
* description and responsible_single are two converters of type <tt>DictTextElement</tt> and can be used to match individual entries in the YAML header contained in the markdown file. | *description and responsible_single are two converters of type <tt>DictTextElement</tt> and can be used to match individual entries in the YAML header contained in the markdown file. | ||
There are many more types of standard converters included in the LinkAhead crawler. Examples include converters for interpreting tabular data (in Excel or | There are many more types of standard converters included in the LinkAhead crawler. Examples include converters for interpreting tabular data (in Excel or .csv format), JSON<ref>{{Cite journal |last=Pezoa |first=Felipe |last2=Reutter |first2=Juan L. |last3=Suarez |first3=Fernando |last4=Ugarte |first4=Martín |last5=Vrgoč |first5=Domagoj |date=2016-04-11 |title=Foundations of JSON Schema |url=https://dl.acm.org/doi/10.1145/2872427.2883029 |journal=Proceedings of the 25th International Conference on World Wide Web |language=en |publisher=International World Wide Web Conferences Steering Committee |place=Montréal Québec Canada |pages=263–273 |doi=10.1145/2872427.2883029 |isbn=978-1-4503-4143-1}}</ref><ref>{{Cite web |last=Internet Engineering Task Force (IETF) |date=21 January 2020 |title=The JavaScript Object Notation (JSON) Data Interchange Format - RFC 7159 |work=Datatracker |url=https://datatracker.ietf.org/doc/rfc7159/ |accessdate=10 August 2023}}</ref> files, or HDF5<ref name=":6" /><ref name=":7" /> files. Custom converters can be created in [[Python (programming language)|Python]] using the LinkAhead crawler Python package. There is a community repository online where community extensions are collected and maintained (see Appendix C). | ||
====Variables==== | ====Variables==== | ||
| Line 213: | Line 212: | ||
====Scanner==== | ====Scanner==== | ||
We call the subroutines that gather information from the file system by applying the crawler definition to a file hierarchy during the process of scanning. The corresponding module of the crawler software is called <tt>Scanner</tt>. The result of a scanning process is a list of LinkAhead <tt>Records</tt>, where the values of the <tt>Properties</tt> are set to the information found. In the example shown in Figure 1, the list will contain at least two <tt>Project Records</tt> with names “ElementaryCharge” and “SpeedOfLight”; three <tt>Experiment Records</ | We call the subroutines that gather information from the file system by applying the crawler definition to a file hierarchy during the process of scanning. The corresponding module of the crawler software is called <tt>Scanner</tt>. The result of a scanning process is a list of LinkAhead <tt>Records</tt>, where the values of the <tt>Properties</tt> are set to the information found. In the example shown in Figure 1, the list will contain at least two <tt>Project Records</tt> with names “ElementaryCharge” and “SpeedOfLight”; three <tt>Experiment Records</tt> with dates “2020-01-01,” “2020-01-02,” and “2020-01-03”; and one <tt>Person Record</tt> for “Florian Spreckelsen.” The remaining steps in the synchronization procedure are to split this list into a list of <tt>Records</tt> that need to be newly inserted and a list of <tt>Records</tt> that need an update. The next subsection describes how the updates are distinguished from the insert operations. Then the subsequent subsection describes the final process of carrying out the transactions. | ||
===Identifiables=== | ===Identifiables=== | ||
| Line 220: | Line 219: | ||
Using the example from Figure 1, we can claim that each <tt>Person</tt> in our RDMS can be uniquely identified by providing a “firstName” and a “lastName.” It is important to point out that the definition of identities for <tt>RecordTypes</tt> can vary highly depending on the usage scenario and environment. There might, of course, be many cases where this example definition is not sufficient, because people can have the same first and last names. | Using the example from Figure 1, we can claim that each <tt>Person</tt> in our RDMS can be uniquely identified by providing a “firstName” and a “lastName.” It is important to point out that the definition of identities for <tt>RecordTypes</tt> can vary highly depending on the usage scenario and environment. There might, of course, be many cases where this example definition is not sufficient, because people can have the same first and last names. | ||
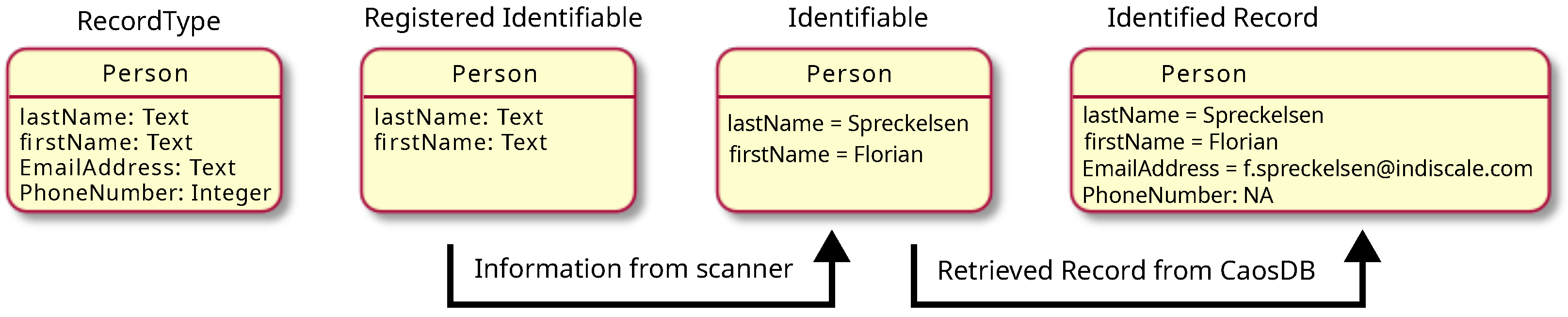
In this case, we would define the identity of <tt>RecordType Person</tt> by declaring that the set of the properties “firstName” and “lastName” is the <tt>Registered Identifiable</tt> for <tt>RecordType Person</tt>. We use the term “Identifiable” here, because we want to avoid confusions with the concept of “unique keys,” which shares some similarities, but are also different in some important aspects. Furthermore, the RDMS LinkAhead also makes use of unique keys as part of its usage of a RDBMS in its back-end. During scanning, <tt>Identifiables</tt> are filled with the necessary information. In our example from Figure 1, “firstName” will be set to “Florian” and “lastName” to “Spreckelsen.” We refer to this entity as the <tt>Identifiable</tt>. Subsequently, this entity is used to check whether a <tt>Record</tt> with these property values already exists in LinkAhead. If there is no such <tt>Record</tt>, a new <tt>Record</tt> is inserted. This <tt>Record</tt> can of course have much more information in the form of properties attached to it, like an “emailAddress”, which is not part of the <tt>Identifiable</tt>. If such a <tt>Record</tt> already exists in LinkAhead, it is retrieved. We refer to the retrieved entity as <tt>Identified Record</tt>. This entity will then be updated as described prior. The terminology, which we introduce here, is summarized in Figure 4. | |||
[[File:Fig4 Wörden Data24 9-2.png|800px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 4.''' Terminology used in the context of defining identity for <tt>Records</tt>. For each <tt>RecordType</tt> that is used in the synchronization procedure, its identity needs to be defined in what we call the <tt>Registered Identifiable</tt>. The scanner will fill the values of the properties of the <tt>Registered Identifiable</tt> and, in that process, create an <tt>Identifiable</tt>. This <tt>Identifiable</tt> can be used by the crawler to run a query on LinkAhead. If a <tt>Record</tt> matching the properties of the <tt>Identifiable</tt> already exists in LinkAhead, it will be retrieved and used to update the <tt>Record</tt>. Otherwise, a new <tt>Record</tt> will be inserted.</blockquote> | |||
|- | |||
|} | |||
|} | |||
To summarize the concept of <tt>Identifiables</tt>: | |||
*An <tt>Identifiable</tt> is a set of properties that is taken from the file system layout or the file contents, which allow for uniquely identifying the corresponding object in the RDMS; | |||
*The set of properties is tied to a specific <tt>RecordType</tt>; | |||
*They may or may not contain references to other objects within the RDMS; and | |||
*Records can of course contain much more information than what is contained in the <tt>Identifiable</tt>. This information is stored in other <tt>Properties</tt>, which are not part of the <tt>Identifiable</tt>. | |||
====Ambiguously defined <tt>Identifiables</tt>==== | |||
Checking the existence of an <tt>Identifiable</tt> in LinkAhead is achieved using a query. If this query returns either zero or exactly one entity, <tt>Identifiables</tt> can be uniquely identified. In case the procedure returns two or more entities, this indicates that the <tt>Identifiable</tt> is not designed properly and that the provided information is not sufficient to uniquely identify an entity. One example might be that there are in fact two people with the same “firstName” and “lastName” in the RDMS. In this case, the <tt>Registered Identifiable</tt> needs to be adapted. The next subsection discusses the design of <tt>Identifiables</tt>. | |||
====Proper design of <tt>Identifiables</tt>==== | |||
The general rule for designing <tt>Identifiables</tt> should be to register the minimum amount of information that needs to be checked to uniquely identify a <tt>Record</tt> of the corresponding type. If too much (unneeded) information is included, it is no longer possible to update parts of the <tt>Record</tt> using information from the file system, as small changes will already lead to a <tt>Record</tt> with a different identity. If, on the other hand, too little information is included in the <tt>Identifiable</tt>, the <tt>Records</tt> might be ambiguously defined, or might become ambiguous in the future. This issue is discussed in the previous paragraph. | |||
To provide an example, we discuss a hypothetical <tt>Record</tt> of <tt>RecordType Experiment</tt> that stores information about a hypothetical experiment carried out in a work group with multiple laboratories. Let us assume that there is only equipment available for one experiment per laboratory and that the maximum number of experiments is one per day. A reasonable set of minimum information for the identifiable could be the date of the experiment and the room number of the laboratory, as it uniquely identifies each experiment. Additional properties, like name of the experimenter, should not be added to the <tt>Identifiable</tt>, as it would reduce flexibility in, e.g., corrections or updates of the respective <tt>Records</tt>. | |||
===Inserts and updates=== | |||
The final step in the crawling procedure is the propagation of the actual transactions into the RDMS. This is illustrated in Figure 2 as Step 3. As described earlier, <tt>Identifiables</tt> can be used to separate the list of <tt>Records</tt>, which were created by the scanner in Step 1, into a list of new <tt>Records</tt> and a list of changed <tt>Records</tt>. The list of new <tt>Records</tt> can just be inserted into the RDMS. For the list of changed <tt>Records</tt>, each entity is retrieved from the server first. Each of these <tt>Records</tt> is then overwritten with the new version of the <tt>Record</tt>, as it was generated by the scanner. Afterwards, the <tt>Records</tt> are updated in the RDMS. | |||
==Discussion== | |||
In Section 2, we presented a concept for data integration that allowed for using the file system simultaneously with an RDMS for managing heterogeneous scientific data using a modularized crawler. It was discussed that the YAML crawler definitions provided a standardized configuration that could be adapted to very different use cases. | |||
In this section, we discuss some important design decisions, benefits of the approach, and limitations. | |||
===YAML specifications as an abstraction layer for data integration=== | |||
In Figure 1, our approach to map the data found on the file system to a semantic data model was presented along with a formalized syntax in YAML. This syntax was designed to be an abstraction of common data integration tasks, like matching directories, files, and their contents, but to still be flexible enough to allow for the integration of very complex data or very rare file formats. | |||
One advantage of using an abstraction layer that is a compromise between very specific source code for data integration and highly standardized routines is that crawler specifications can be re-used in a greater variety of scenarios. Because the hierarchical structure of the YAML crawler specification corresponds to the hierarchical structure of file systems and file contents, it is much simpler to identify similarities between different file structures, than it would be with a plain data integration source code. Furthermore, the crawler specifications are machine-readable and, therefore, open the possibility for much more complex applications. | |||
===Primary source of information=== | |||
Our update procedure synchronizes data from the file system into the RDMS uni-directionally, so we can state that the file system is the single source of truth. Another possibility would have been to implement merges between entities generated from the file system information and entities present in the RDMS. This would have allowed for editing entities simultaneously on the file system and in the RDMS, and to obtain a merged version after the crawling procedure. However, we decided against this and chose a single source of information approach. In practice, merges can become really complicated, and having a clearly defined source of information makes the procedure much more transparent and predictable to users. Furthermore, we found that use cases involving simultaneous edits of entities on the file system and in the RDMS are rather rare, so we decided against adding this additional complexity into our software. | |||
We considered it best practice to not edit entities generated by the crawler in the RDMS directly, but to use references to these entities instead. This could be enforced by LinkAhead by automatically setting the entities generated by the crawler as read-only for other users. LinkAhead entities are protected against accidental data loss using versioning of entities. | |||
===Documentation of data structures and file hierarchies=== | |||
The approach we presented here purposefully relies on a manual definition of the semantic data model and a careful definition of the synchronization rules. Sometimes, it has been criticized that this can involve a lot of work. A frequent suggestion is to instead apply [[machine learning]] (ML) techniques or [[artificial intelligence]] (AI) methods to organize data and make it more findable. However, it is important to highlight that the manual design and documentation of file structures and file hierarchies actually has several beneficial side effects. | |||
One of the main goals of managing and organizing data is to enable researchers to better understand, find, and re-use their data.<ref name=":0" /> A semantic data model created by hand results in researchers understanding their data. Data organized by AI is very likely not represented in a way that is easily understood by the researcher and it probably does not incorporate the same meaning. The process of designing the data models and rules for synchronization is a creative process that leads to optimized research workflows. In our experience, researchers highly benefit from the process of structuring their own data management. | |||
One disadvantage of ML methods is that many of them rely on large amounts of training data for being accurate and efficient. Often these data are not available, and therefore a manual step that is likely to be equally time-consuming to the design of the data model and synchronization rules is necessary. | |||
As a practical outcome, the data documentation created in the form of crawler definitions can be used to create data management plans, which are nowadays an important requirement for institutions and funding agencies. Although we think that the design of the process should be, in part, manual, integrating AI in the form of assistants is possible. An example for such an assistant could be an algorithm that generates a suggested crawler definition based on existing data, which then could be corrected and expanded on by the researcher. | |||
===Limitations=== | |||
====Deletion of files==== | |||
One important limitation of the approach presented here is that deletions on the file system are not directly mapped to the RDMS, i.e., the records stemming from deleted files will persist in the RDMS and have to be deleted manually. This design decision was intended as it allows for complex distributed workflows. One example is that of two researchers from the same work group work on two different projects with independent file structures. Using our approach, it is possible to run the crawler on two independent file trees and thereby update different parts of the RDMS, without having to synchronize the file systems before. | |||
Future implementations of the software could make use of file system monitoring to implement proper detection of deleted files. Another possibility would be to signal the deletion of files and folders by special files (e.g., special names or contents) that trigger an automatic deletion by the crawler. | |||
====Bi-directional synchronization==== | |||
A natural extension of the concepts presented here would be a crawler that allows for bi-directional synchronization. In addition to inserts and updates that are propagated from the file structure to the RDMS, changes in RDMS would also be detected and propagated to the file system, leading to the creation and updating of files. While some parts of these procedure, like identifying changes in the RDMS, can be implemented in a straight-forward way, the mapping of information to existing file trees can be considered quite complex and raises several questions. While the software in its current form needs only read-only access to the file system, in a bi-directional scenario, read–write–access is required, so that more care has to be taken to protect the users from unwanted data loss. | |||
==Conclusions== | |||
In this article, we present a structured approach for data integration from file systems into RDMS. Using a simplified example, we have shown how this concept is applied practically, and we have published an open-source software framework as one implementation of this concept (see Appendix A). In multiple active data management projects (some of them are mentioned in the Appendix B), we found that this mixture of a standardized definition of the data integration with the possibility to extend them with flexible custom code allows for a rapid development of data integration tools and facilitates the re-use of data integration modules. In these projects, we experienced that using the presented standardized YAML format highly facilitated the integration of data sets, especially in cases where data were stored in non-standardized directory layouts. We were also able to re-use our <tt>CFoods</tt>, which only required minor adaptions when transferring them to a different context. To foster re-usability, we set up a community repository for <tt>CFoods</tt> (see Appendix C). | |||
Our current focus for the software is on transferring the concept to multiple different use cases in order to make the crawler more robust and to identify the limitations and usability issues. Furthermore, the bi-directional synchronization concern mentioned prior and an assistant for creating <tt>CFoods</tt> that is based on ML are currently under investigation as possible next features. | |||
==Appendices== | ==Appendices== | ||
| Line 226: | Line 293: | ||
The following software projects can be used to implement the workflows described in the article: | The following software projects can be used to implement the workflows described in the article: | ||
* Repository of the LinkAhead open-source project: https://gitlab.com/linkahead | *Repository of the LinkAhead open-source project: https://gitlab.com/linkahead | ||
* Repository of the LinkAhead crawler: https://gitlab.com/linkahead/linkahead-crawler | *Repository of the LinkAhead crawler: https://gitlab.com/linkahead/linkahead-crawler | ||
The installation procedures for LinkAhead and the crawler framework are provided in their respective repositories. Also a Docker container is available for the instant deployment of LinkAhead. | The installation procedures for LinkAhead and the crawler framework are provided in their respective repositories. Also a Docker container is available for the instant deployment of LinkAhead. | ||
| Line 239: | Line 306: | ||
===Appendix D. Software documentation=== | ===Appendix D. Software documentation=== | ||
The official documentation for LinkAhead, including an installation guide, can be found at https://docs.indiscale.com. The documentation for the crawler framework that is presented in this article can be found at https://docs.indiscale.com/caosdb-crawler/. | The official documentation for LinkAhead, including an installation guide, can be found at https://docs.indiscale.com. The documentation for the crawler framework that is presented in this article can be found at https://docs.indiscale.com/caosdb-crawler/. | ||
==Abbreviations, acronyms, and initialisms== | |||
*'''AI''': artificial intelligence | |||
*'''API''': application programming interface | |||
*'''DBMS''': database management system | |||
*'''ELN''': electronic laboratory notebook | |||
*'''ETL''': extract, transform, and load | |||
*'''FAIR''': findable, accessible, interoperable, and reusable | |||
*'''FDO''': FAIR Digital Object | |||
*'''GUI''': graphical user interface | |||
*'''IT''': information technology | |||
*'''ML''': machine learning | |||
*'''PID''': persistent identifier | |||
*'''RDBMS''': relational database management system | |||
*'''RDMS''': research data management system | |||
==Acknowledgements== | |||
The authors thank Birte Hemmelskamp-Pfeiffer for providing information on https://dataportal.leibniz-zmt.de. | |||
===Author contributions=== | |||
Conceptualization, A.S., H.t.W. and F.S.; software, A.S., H.t.W. and F.S.; resources, A.S., H.t.W., U.P., S.L. and F.S.; writing—original draft preparation, A.S., H.t.W. and F.S.; writing—review and editing, A.S., H.t.W., U.P., S.L. and F.S.; supervision, A.S. and H.t.W.; project administration, H.t.W. and S.L.; funding acquisition, A.S., U.P. and S.L. All authors have read and agreed to the published version of the manuscript. | |||
===Funding=== | |||
A.S., U.P., and S.L. acknowledge financial support from the Volkswagen Stiftung within the framework of the project “Global Carbon Cycling and Complex Molecular Patterns in Aquatic Systems: Integrated Analyses Powered by Semantic Data Management”. S.L. acknowledges financial support from the DZHK and DFG SFB 1002 Modulary Units in Heart Failure, and the Else Kröner-Fresenius Foundation (EKFS). | |||
===Conflicts of interest=== | |||
A.S. and H.t.W. are co-founders of IndiScale GmbH, a company providing commercial services for LinkAhead. Furthermore, H.t.W. and F.S. are employees of IndiScale GmbH. A.S. is external scientific advisor for IndiScale GmbH. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. | |||
==References== | ==References== | ||
Latest revision as of 17:50, 11 June 2024
| Full article title | Mapping hierarchical file structures to semantic data models for efficient data integration into research data management systems |
|---|---|
| Journal | Data |
| Author(s) | tom Wörden, Henrik; Spreckelsen, Florian; Luther, Stefan; Parlitz, Ulrich; Schlemmer, Alexander |
| Author affiliation(s) | Indiscale GmbH, Max Planck Institute for Dynamics and Self-Organization, Georg-August-Universität, German Center for Cardiovascular Research (DZHK) Göttingen, University Medical Center Göttingen |
| Primary contact | Email: alexander dot schlemmer at ds dot mpg dot de |
| Editors | Sedig, Kamran |
| Year published | 2024 |
| Volume and issue | 9(2) |
| Page(s) | 24 |
| DOI | 10.3390/data9020024 |
| ISSN | 2306-5729 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.mdpi.com/2306-5729/9/2/24 |
| Download | https://www.mdpi.com/2306-5729/9/2/24/pdf (PDF) |
Abstract
Although other methods exist to store and manage data using modern information technology (IT), the standard solution is file systems. Therefore, maintaining well-organized file structures and file system layouts can be key to a sustainable research data management infrastructure. However, file structures alone lack several important capabilities for FAIR (findable, accessible, interoperable, and reusable) data management, the two most significant being insufficient visualization of data and inadequate possibilities for searching and obtaining an overview. Research data management systems (RDMSs) can fill this gap, but many do not support the simultaneous use of the file system and RDMS. This simultaneous use can have many benefits, but keeping data in an RDMS in synchrony with the file structure is challenging.
Here, we present concepts that allow for keeping file structures and semantic data models (found in RDMSs) synchronous. Furthermore, we propose a specification in YAML format that allows for a structured and extensible declaration and implementation of a mapping between the file system and data models used in semantic research data management. Implementing these concepts will facilitate the re-use of specifications for multiple use cases. Furthermore, the specification can serve as a machine-readable and, at the same time, human-readable documentation of specific file system structures. We demonstrate our work using the open-source RDMS LinkAhead (previously named “CaosDB”).
Keywords: research data management, FAIR, file structure, file crawler, metadata, semantic data model
Introduction
Data management for research is part of an active transformation in science, with effective management required in order to meet the needs of increasing amounts of complex data. Furthermore, the FAIR guiding principles[1] for scientific data—which are an elementary part of numerous data management plans, funding guidelines, and data management strategies of research organizations[2][3], requiring that research objects be more findable, accessible, interoperable, and reusable—require scientists to review and enhance their established data management workflows.
One particular focus of this endeavor is the introduction and expansion of research data management systems (RDMSs). These systems help researchers organize their data during the whole data management life cycle, especially by increasing findability and accessibility.[4] Furthermore, semantic data management approaches[5] can increase the reuse and reproducibility of data that are typically organized in file structures. As has been pointed out by Gray et al.[4], one major shortcoming of file systems is the lack of rich metadata features, which additionally limits search options. Typically, RDMSs employ database management systems (DBMSs) to store data and metadata, but the degree to which data is migrated, linked, or synchronized into these systems can vary substantially.
The import of data into an RDMS typically requires the development of data integration procedures that are tied to the specific workflows at hand. While very few standard products exist[6], in practice, mostly custom software written in various programming languages and making use of a high variety of different software packages are used for data integration in scientific environments. There are two main workflows for integrating data into RDMSs: manually inputting data (e.g., using forms[7]) or facilitating the batch import of data sets. The automatic methods often include data import routines for predefined formats, like tables in Excel or .csv format.[8][9] Some systems include plugin systems to allow for a configuration of the data integration process.[10] Sometimes, data files have to be uploaded using a web front-end[11] and are afterwards attached to objects in the RDMSs. In general, developing this kind of software can be considered very costly[6], as it is highly dependent on the specific environment. Data import can still be considered one of the major bottlenecks for the adaption of an RDMS.
There are several advantages to using an RDMS over organization of data in classical file hierarchies. There is a higher flexibility in adding metadata to data sets, while these capabilities are limited for classical file systems. The standardized representation in an RDMS improves the comparability of data sets that possibly originate from different file formats and data representations. Furthermore, semantic information can be seamlessly integrated, possibly using standards like RDF[12] and OWL.[13] The semantic information allows for advanced querying and searching, e.g., using SPARQL.[14] Concepts like linked data[15][16] and FAIR digital objects (FDO[17]) provide overarching concepts for achieving more standardized representations within RDMSs and for publication on the web. Specifically, the FDO concept aims at bundling data sets with a persistent identifier (PID) and its metadata to self-contained units. These units are designed to be machine-actionable and interoperable, so that they have the potential to build complex and distributed data processing infrastructures.[17]
Using file systems and RDMSs simultaneously
Despite the advantages mentioned above, RDMSs have still failed to gain a widespread adoption. One of the key problems in the employment of an RDMS in an active research environment is that a full transition to such a system is very difficult, as most digital scientific workflows are in one or multiple ways dependent on classical hierarchical file systems.[4] Examples include data acquisition and measurement devices, data processing and analysis software, and digitized laboratory notes and material for publications. The complete transition to an RDMS would require developing data integration procedures (e.g., extract, transform, load [ETL][6][18] processes) for every digital workflow in the lab and to provide interfaces for input and output to any other software involved in these workflows.
As files on classical file systems play a crucial role in these workflows, our aim is to develop a robust strategy to use file systems and an RDMS simultaneously. Rather than requiring a full transition to an RDMS, we want to make use of the file system as an interoperability layer between the RDMS and any other file-based workflow in the research environment.
There are two important tasks that need to be solved and that are the main focus of this article:
- There must be a method to keep data and metadata in the RDMS synchronized with data files on the file system. Using that method, the file system can be used as an interoperability layer between the RDMS and other software and workflows. Our approach to solving this issue is discussed in detail in the results section. One key component of the synchronization method is defining the concept of "identity" for data in the RDMS, also discussed in the results section about identifiables.
- The high variety of different data structures found on the file system needs an adaptive and flexible approach for data integration and synchronization into the RDMS. We discuss our solution for this task in the results section concerning YAML, where we present a standardized but highly configurable format for mapping information from files to a semantic data model.
Apart from the main motivation, described above, we have identified several additional advantages of using a conventional folder structure simultaneous to an RDMS: standard tools for managing the files can be used for backup (e.g., rsync), versioning (e.g., git), archiving, and file access (e.g., SSH). Functionality of these tools does not need to be re-implemented in the RDMS. Furthermore, the file system can act as a fallback in cases where the RDMS might become unavailable. This methodology, therefore, increases robustness. As a third advantage, existing workflows relying on storing files in a file system do not need to be changed, while the simultaneous findability within an RDMS is available to users.
The concepts described in this article can be used independent of a specific RDMS software. However, as a proof-of-concept, we implemented the approach as part of the file crawler framework that belongs to the open-source RDMS LinkAhead (recently renamed from CaosDB).[19][20] The crawler framework is released as open-source software under the AGPLv3 license (see Appendix A).
Example data set
We will illustrate the problem of integrating research data using a simplified example that is based on the work of Spreckelsen et al.[21] This example will be used in the results section to demonstrate our data integration concepts. Examples for more complex data integration, e.g., for data sets found in the neurosciences (BIDS[22] and DICOM[23]) and in the geosciences, can be found online (see Appendix B). Although the concept is not restricted to data stored on file systems, in this example we will assume for simplicity that the research data are stored on a standard file system with a well-defined file structure layout:
ExperimentalData/
- 2020_SpeedOfLight/
- 2020-01-01_TimeOfFlight
- README.md
- ...
- README.md
- 2020-01-02_Cavity
- README.md
- ...
- README.md
- 2020-01-01_TimeOfFlight
- 2020-01-03
- 2020-01-03
- README.md
- ...
- README.md
The above listing replicates an example with experimental data from Spreckelsen et al.[21] using a three-level folder structure:
- Level 1 (ExperimentalData) stores rough categories for data, in this data acquired from experimental measurements.
- Level 2 (2020_SpeedOfLight) is the level of project names, grouping data into independent projects.
- Level 3 stores the actual measurement folders, which can also be referred to as “scientific activity” folders in the general case. Each of these folders could have an arbitrary substructure and store the actual experimental data along with a README.md file, containing meta data.
The generic use case of integrating data from file systems involves the following sub tasks:
- Identify the required data for integration into the RDMS. This can possibly involve information contained in the file structure (e.g., file names, path names, or file extensions) or data contained in the contents of the files themselves.
- Define an appropriate (semantic) data model for the desired data.
- Specify the data integration procedure that maps data found on the file system (including data within the files) to the (semantic) data in the RDMS.
A concrete example for this procedure, including a semantic data model, is provided in the results section. As previously state, there are already many use cases that can benefit from the simultaneous use of the file system and RDMS. Therefore, it is important to implement reliable means for identifying and transferring the data not only once, as a single “data import,” but also while allowing for frequent updates of existing or changed data. Such an update might be needed if an error in the raw data has been detected. It can then be corrected on the file system, with the changes needing to be propagated to the RDMS. Another possibility is that data files that are actively worked on have been inserted into the RDMS. A third-party software is used to process these files and, consequently, the information taken from the files has to be frequently updated in the RDMS.
We use the term “synchronization” here to refer to the possible insertion of new data sets and to update existing data sets in the same procedure. To avoid confusion, we want to explicitly note here that we are not referring to bi-directional synchronization. Bi-directional synchronization means that information from RDMS that is not present in the file system can be propagated back to the file system, which is not possible in our current implementation. Although ideas exist to implement bi-directional synchronization in the future, in the current work (and also the current software implementation), we focus on the uni-directional synchronization from the file system to the RDMS. The outlook for adding extensions to bi-directional synchronization will be discussed in the discussion section.
About LinkAhead
LinkAhead was designed as an RDMS mainly targeted at active data analysis. So, in contrast to electronic laboratory notebooks (ELNs), which have a stronger focus on data acquisition and data repositories, which are used to publish data, data in LinkAhead are assumed to be actively worked on by scientists on a regular basis. Its scope for single instances (which are usually operated on-premises) ranges from small work groups to whole research institutes. Independent of any RDMS, data acquisition typically leads to files stored on a file system. The LinkAhead crawler synchronizes data from the file system into the RDMS. LinkAhead provides multiple interfaces for interacting with the data, such as a web-based graphical user interface (GUI) and an application programming interface (API) that can be used for interfacing the RDMS from multiple programming languages. LinkAhead itself is typically not used as a data repository, but the structured and enriched data in LinkAhead serves as a preparation for data publication, and data can be exported from the system and published in data repositories. The semantic data model used by LinkAhead is described in more detail in the next subsection. LinkAhead is open-source software, released under the AGPLv3 license (see Appendix A).
Data models in LinkAhead
The LinkAhead data model is basically an object-oriented representation of data which makes use of four different types of entities: RecordType, Property, Record and File RecordTypes, and Properties. These four entities define the data model, which is later used to store concrete data objects, which are represented by Records. In that respect, RecordTypes and Properties share a lot of similarities with ontologies, but have a restricted set of relations, as described in more detail by Fitschen et al.[19] Files have a special role within LinkAhead as they represent references to actual files on a file system, but allow for linking them to other LinkAhead entities and, e.g., adding custom properties.
Properties are individual pieces of information that have a name, description, optionally a physical unit, and can store a value of a well-defined data type. Properties are attached to RecordTypes and can be marked as “obligatory,” “recommended,” or “suggested.” In case of obligatory Properties, each Record of the respective RecordType is enforced to set the respective Properties. Each Record must have at least one RecordType and RecordTypes can have other RecordTypes as parents. This is known as (multiple) inheritance in object-oriented programming languages.
In Figure 1, an example data model is shown in the right column in a UML-like diagram. There are three RecordTypes (Project, Person, and Experiment), each with a small set of Properties (e.g., an integer Property called “year” or a Property referring to records of type Person called “responsible”). The red lines with a diamond show references between RecordTypes, i.e., where RecordTypes are used as Properties in other RecordTypes.
|
Results
We solved the issues described prior about using file systems and RDMSs simultaneously by using a modular crawler system, which is discussed extensively in this section. The main task of the crawler is to automatically synchronize information found in the file system to the semantic RDMS. The modularity of the crawler is achieved by providing a flexible configuration of synchronization and mapping rules in a human- and machine-readable YAML format. Using these configuration files (which we will refer to as CFoods), it is possible to adapt the crawler to heterogeneous use cases. (These crawler definitions will be described a bit later in this section.) We assumed that a semantic data model that is appropriate for the data structures had been created. As an example, we discuss the data model that is shown in Figure 1 in the right column.
In order to be able to synchronize information from the file system with the RDMS, a specification of the identity of the objects is needed. This will allow us to check which objects are already present in the RDMS and, therefore, need an update instead of an insert operation. Our concept, which is similar to unique keys in relational database management systems (RDBMSs), will be discussed later in this section. Our implementation of the procedure, the LinkAhead crawler, makes use of these concepts in order to integrate data into the RDMS. Based on the example we introduced in the prior section, we will illustrate in the following subsections how the information from the file system will be mapped onto semantic data in the RDMS.
The LinkAhead crawler
Figure 2 provides an overview of the complete data integration procedure with the LinkAhead crawler. In Step 1, the scanner uses the YAML crawler definition to match converters to a given file system tree. From the information that is matched, a list of LinkAhead Records with Properties is created. In Step 2, the crawler checks which of these Records are already contained in LinkAhead in order to separate the list of Records into a list of new Records and a list of changed Records. In order to complete this check, it makes use of a given definition of Identifiables. In Step 3 both lists are synchronized with the LinkAhead server, i.e., all Records from the list of new Records are inserted into LinkAhead and all Records from the list of changed Records are updated. The data model is needed to write a valid YAML crawler definition and to create the definition of Identifiables. Furthermore, it is used by the LinkAhead server directly.
|
Mapping files and layouts into a data model
Suppose we have a hierarchical structure of some kind that contains certain information and we want to map this information onto our object-oriented data model. The typical example for the hierarchical structure would be a folder structure with files, but hierarchical data formats like HDF5[24][25] files (or a mixture of both) would also fit the use case.
The prior Figure 1 illustrates what such a mapping could look like in practice using the prior stated file structure:
- ExperimentalData contains one subfolder 2020_SpeedOfLight, storing all data from this experimental series. The experimental series is represented in a RecordType called Project. The Properties “year” (2020) and “identifier” (SpeedOfLight) can be directly filled from the directory name.
- Each experiment of the series has its dedicated subfolder, so one Record of type Experiment will be created for each one. The association to its Project, which is implicitly clear in the folder hierarchy, can be mapped to a reference to the Project Record created in the previous step. Again, the Property “date” can be set from the directory name.
- Each experiment contains a file called README.md storing text and metadata about the experiment, according to the described standard. In this case, the “description” Property for the Experiment Record created in the previous step will be set from the corresponding YAML value. Furthermore, a Person Record with Properties "firstName = Florian" and "lastName = Spreckelsen" will be created. Finally the Person Record will be set as the value for the property “responsible” of the Experiment Record.
YAML definitions
Figure 3 illustrates the modular design of the crawler. Data acquisition and other scientific activities can lead to heterogeneous file structures involving very different data formats. The crawler uses crawler specifications (CFoods) that define how these structures are interpreted and synchronized with LinkAhead. We explain this description using an example of CFood in this section. CFood is designed to make use of the example file structure discussed in the introduction.
|
As we pointed out in the introduction, one major challenge is allowing for the adaptation of data integration to very different and potentially very heterogeneous use cases. Therefore, we designed a special syntax in YAML format for configuring the mapping which were just described in a machine-readable form. We call this description of rules for the crawler the crawler definition or CFood. For the example file structure, the corresponding YAML file would look like this:
ExperimentalData_Dir:
- type: Directory
- match: ExperimentalData
- subtree:
- Project_Dir:
- type: Directory
- match: (?P<year>[0-9]{4,4})_(?P<name>.*)
- records:
- Project:
- year: $year
- name: $name
- year: $year
- Project:
- subtree:
- Experiment_Dir:
- type: Directory
- match: >- (?P<date>[0-9]{4,4}_[0-9]{2,2}_[0-9]{2,2})(_(?P<identifier>.*))?
- records:
- Experiment:
- Project: $Project
- date: $date
- identifier: $identifier
- Project: $Project
- Experiment:
- subtree:
- Readme_File:
- type: MarkdownFile
- match: README.md
- subtree:
- description:
- type: DictTextElement
- match_value: (?P<description>.*)
- match_name: description
- records:
- Experiment:
- description: $description
- description: $description
- Experiment:
- type: DictTextElement
- responsible_single:
- type: DictTextElement
- match_name: responsible
- match_value: ((?P<first_name>.+) )?(?P<last_name>.+)
- records:
- Person:
- first_name: $first_name
- last_name: $last_name
- first_name: $first_name
- Experiment:
- Person: $Person
- Person:
- type: DictTextElement
- description:
- type: MarkdownFile
- Readme_File:
- type: Directory
- Experiment_Dir:
- type: Directory
- Project_Dir:
The YAML structure mimics the hierarchical structure of a file tree. The crawler operates by successively matching converters to files, folders, and possibly other StructureElements (A StructureElement specifies any piece of information that is derived from some part of the file structure and that can be matched by a converter.) If a converter matches, Records are created corresponding to the entries under the Records in the crawler definition. Afterwards, the crawler proceeds by processing sub-elements, like sub-folders, using the converters defined under the subtree.
In the given example, the following converters can be found:
- ExperimentalData_Dir, Project_Dir, Experiment_Dir, which are all of type Directory. These converters match names of folders against the regular expression given by match. When matching, converters of this type yield sub-folders and -files for processing of converters in the subtree section.
- Readme_File is of type MarkdownFile and allows for processing of the contents of the YAML header by converters given in the subtree section.
- description and responsible_single are two converters of type DictTextElement and can be used to match individual entries in the YAML header contained in the markdown file.
There are many more types of standard converters included in the LinkAhead crawler. Examples include converters for interpreting tabular data (in Excel or .csv format), JSON[27][28] files, or HDF5[24][25] files. Custom converters can be created in Python using the LinkAhead crawler Python package. There is a community repository online where community extensions are collected and maintained (see Appendix C).
Variables
Variables begin with a dollar sign and can be used in multiple occasions within the CFood, e.g., for setting properties of Records. These variables and their values are set during multiple operations within the crawling procedure; variables that are part of a matched regular expression will be available. For example, $year is set from the match entry of the converter Project_Dir. Record definitions within the crawler definition will be available as variables, e.g., the records section of Project_Dir creates a variable $Project, which will later be used as the value of a Property of Record Experiment created in the records section of Experiment_Dir. More details about the syntax are made available in the official documentation of the LinkAhead crawler software (see Appendix D).
Scanner
We call the subroutines that gather information from the file system by applying the crawler definition to a file hierarchy during the process of scanning. The corresponding module of the crawler software is called Scanner. The result of a scanning process is a list of LinkAhead Records, where the values of the Properties are set to the information found. In the example shown in Figure 1, the list will contain at least two Project Records with names “ElementaryCharge” and “SpeedOfLight”; three Experiment Records with dates “2020-01-01,” “2020-01-02,” and “2020-01-03”; and one Person Record for “Florian Spreckelsen.” The remaining steps in the synchronization procedure are to split this list into a list of Records that need to be newly inserted and a list of Records that need an update. The next subsection describes how the updates are distinguished from the insert operations. Then the subsequent subsection describes the final process of carrying out the transactions.
Identifiables
In order to determine which of the Records that have been generated during the scanning procedure are already present in the RDMS, the crawler needs information on how to determine the identity of Records. For this purpose, we implemented a concept that is similar to unique keys, which are used in the context of RDBMSs. For each RecordType, we define a set of Properties that can be used to uniquely identify a single Record.
Using the example from Figure 1, we can claim that each Person in our RDMS can be uniquely identified by providing a “firstName” and a “lastName.” It is important to point out that the definition of identities for RecordTypes can vary highly depending on the usage scenario and environment. There might, of course, be many cases where this example definition is not sufficient, because people can have the same first and last names.
In this case, we would define the identity of RecordType Person by declaring that the set of the properties “firstName” and “lastName” is the Registered Identifiable for RecordType Person. We use the term “Identifiable” here, because we want to avoid confusions with the concept of “unique keys,” which shares some similarities, but are also different in some important aspects. Furthermore, the RDMS LinkAhead also makes use of unique keys as part of its usage of a RDBMS in its back-end. During scanning, Identifiables are filled with the necessary information. In our example from Figure 1, “firstName” will be set to “Florian” and “lastName” to “Spreckelsen.” We refer to this entity as the Identifiable. Subsequently, this entity is used to check whether a Record with these property values already exists in LinkAhead. If there is no such Record, a new Record is inserted. This Record can of course have much more information in the form of properties attached to it, like an “emailAddress”, which is not part of the Identifiable. If such a Record already exists in LinkAhead, it is retrieved. We refer to the retrieved entity as Identified Record. This entity will then be updated as described prior. The terminology, which we introduce here, is summarized in Figure 4.
|
To summarize the concept of Identifiables:
- An Identifiable is a set of properties that is taken from the file system layout or the file contents, which allow for uniquely identifying the corresponding object in the RDMS;
- The set of properties is tied to a specific RecordType;
- They may or may not contain references to other objects within the RDMS; and
- Records can of course contain much more information than what is contained in the Identifiable. This information is stored in other Properties, which are not part of the Identifiable.
Ambiguously defined Identifiables
Checking the existence of an Identifiable in LinkAhead is achieved using a query. If this query returns either zero or exactly one entity, Identifiables can be uniquely identified. In case the procedure returns two or more entities, this indicates that the Identifiable is not designed properly and that the provided information is not sufficient to uniquely identify an entity. One example might be that there are in fact two people with the same “firstName” and “lastName” in the RDMS. In this case, the Registered Identifiable needs to be adapted. The next subsection discusses the design of Identifiables.
Proper design of Identifiables
The general rule for designing Identifiables should be to register the minimum amount of information that needs to be checked to uniquely identify a Record of the corresponding type. If too much (unneeded) information is included, it is no longer possible to update parts of the Record using information from the file system, as small changes will already lead to a Record with a different identity. If, on the other hand, too little information is included in the Identifiable, the Records might be ambiguously defined, or might become ambiguous in the future. This issue is discussed in the previous paragraph.
To provide an example, we discuss a hypothetical Record of RecordType Experiment that stores information about a hypothetical experiment carried out in a work group with multiple laboratories. Let us assume that there is only equipment available for one experiment per laboratory and that the maximum number of experiments is one per day. A reasonable set of minimum information for the identifiable could be the date of the experiment and the room number of the laboratory, as it uniquely identifies each experiment. Additional properties, like name of the experimenter, should not be added to the Identifiable, as it would reduce flexibility in, e.g., corrections or updates of the respective Records.
Inserts and updates
The final step in the crawling procedure is the propagation of the actual transactions into the RDMS. This is illustrated in Figure 2 as Step 3. As described earlier, Identifiables can be used to separate the list of Records, which were created by the scanner in Step 1, into a list of new Records and a list of changed Records. The list of new Records can just be inserted into the RDMS. For the list of changed Records, each entity is retrieved from the server first. Each of these Records is then overwritten with the new version of the Record, as it was generated by the scanner. Afterwards, the Records are updated in the RDMS.
Discussion
In Section 2, we presented a concept for data integration that allowed for using the file system simultaneously with an RDMS for managing heterogeneous scientific data using a modularized crawler. It was discussed that the YAML crawler definitions provided a standardized configuration that could be adapted to very different use cases.
In this section, we discuss some important design decisions, benefits of the approach, and limitations.
YAML specifications as an abstraction layer for data integration
In Figure 1, our approach to map the data found on the file system to a semantic data model was presented along with a formalized syntax in YAML. This syntax was designed to be an abstraction of common data integration tasks, like matching directories, files, and their contents, but to still be flexible enough to allow for the integration of very complex data or very rare file formats.
One advantage of using an abstraction layer that is a compromise between very specific source code for data integration and highly standardized routines is that crawler specifications can be re-used in a greater variety of scenarios. Because the hierarchical structure of the YAML crawler specification corresponds to the hierarchical structure of file systems and file contents, it is much simpler to identify similarities between different file structures, than it would be with a plain data integration source code. Furthermore, the crawler specifications are machine-readable and, therefore, open the possibility for much more complex applications.
Primary source of information
Our update procedure synchronizes data from the file system into the RDMS uni-directionally, so we can state that the file system is the single source of truth. Another possibility would have been to implement merges between entities generated from the file system information and entities present in the RDMS. This would have allowed for editing entities simultaneously on the file system and in the RDMS, and to obtain a merged version after the crawling procedure. However, we decided against this and chose a single source of information approach. In practice, merges can become really complicated, and having a clearly defined source of information makes the procedure much more transparent and predictable to users. Furthermore, we found that use cases involving simultaneous edits of entities on the file system and in the RDMS are rather rare, so we decided against adding this additional complexity into our software.
We considered it best practice to not edit entities generated by the crawler in the RDMS directly, but to use references to these entities instead. This could be enforced by LinkAhead by automatically setting the entities generated by the crawler as read-only for other users. LinkAhead entities are protected against accidental data loss using versioning of entities.
Documentation of data structures and file hierarchies
The approach we presented here purposefully relies on a manual definition of the semantic data model and a careful definition of the synchronization rules. Sometimes, it has been criticized that this can involve a lot of work. A frequent suggestion is to instead apply machine learning (ML) techniques or artificial intelligence (AI) methods to organize data and make it more findable. However, it is important to highlight that the manual design and documentation of file structures and file hierarchies actually has several beneficial side effects.
One of the main goals of managing and organizing data is to enable researchers to better understand, find, and re-use their data.[1] A semantic data model created by hand results in researchers understanding their data. Data organized by AI is very likely not represented in a way that is easily understood by the researcher and it probably does not incorporate the same meaning. The process of designing the data models and rules for synchronization is a creative process that leads to optimized research workflows. In our experience, researchers highly benefit from the process of structuring their own data management.
One disadvantage of ML methods is that many of them rely on large amounts of training data for being accurate and efficient. Often these data are not available, and therefore a manual step that is likely to be equally time-consuming to the design of the data model and synchronization rules is necessary.
As a practical outcome, the data documentation created in the form of crawler definitions can be used to create data management plans, which are nowadays an important requirement for institutions and funding agencies. Although we think that the design of the process should be, in part, manual, integrating AI in the form of assistants is possible. An example for such an assistant could be an algorithm that generates a suggested crawler definition based on existing data, which then could be corrected and expanded on by the researcher.
Limitations
Deletion of files
One important limitation of the approach presented here is that deletions on the file system are not directly mapped to the RDMS, i.e., the records stemming from deleted files will persist in the RDMS and have to be deleted manually. This design decision was intended as it allows for complex distributed workflows. One example is that of two researchers from the same work group work on two different projects with independent file structures. Using our approach, it is possible to run the crawler on two independent file trees and thereby update different parts of the RDMS, without having to synchronize the file systems before.
Future implementations of the software could make use of file system monitoring to implement proper detection of deleted files. Another possibility would be to signal the deletion of files and folders by special files (e.g., special names or contents) that trigger an automatic deletion by the crawler.
Bi-directional synchronization
A natural extension of the concepts presented here would be a crawler that allows for bi-directional synchronization. In addition to inserts and updates that are propagated from the file structure to the RDMS, changes in RDMS would also be detected and propagated to the file system, leading to the creation and updating of files. While some parts of these procedure, like identifying changes in the RDMS, can be implemented in a straight-forward way, the mapping of information to existing file trees can be considered quite complex and raises several questions. While the software in its current form needs only read-only access to the file system, in a bi-directional scenario, read–write–access is required, so that more care has to be taken to protect the users from unwanted data loss.
Conclusions
In this article, we present a structured approach for data integration from file systems into RDMS. Using a simplified example, we have shown how this concept is applied practically, and we have published an open-source software framework as one implementation of this concept (see Appendix A). In multiple active data management projects (some of them are mentioned in the Appendix B), we found that this mixture of a standardized definition of the data integration with the possibility to extend them with flexible custom code allows for a rapid development of data integration tools and facilitates the re-use of data integration modules. In these projects, we experienced that using the presented standardized YAML format highly facilitated the integration of data sets, especially in cases where data were stored in non-standardized directory layouts. We were also able to re-use our CFoods, which only required minor adaptions when transferring them to a different context. To foster re-usability, we set up a community repository for CFoods (see Appendix C).
Our current focus for the software is on transferring the concept to multiple different use cases in order to make the crawler more robust and to identify the limitations and usability issues. Furthermore, the bi-directional synchronization concern mentioned prior and an assistant for creating CFoods that is based on ML are currently under investigation as possible next features.
Appendices
Appendix A. Supporting software
The following software projects can be used to implement the workflows described in the article:
- Repository of the LinkAhead open-source project: https://gitlab.com/linkahead
- Repository of the LinkAhead crawler: https://gitlab.com/linkahead/linkahead-crawler
The installation procedures for LinkAhead and the crawler framework are provided in their respective repositories. Also a Docker container is available for the instant deployment of LinkAhead.
Appendix B. Example crawlers
There is a documented example available online ( at https://gitlab.com/linkahead/crawler-extensions/documented-crawler-example) that demonstrates the application of the crawler to example data. This can also be used as a template for the development of custom crawlers. We are currently aware of one public instance of LinkAhead, which makes use of a complex crawler based on the crawler framework described in this article. It is provided by ZMT-Leibniz Centre for Tropical Marine Research and can be accessed online at https://dataportal.leibniz-zmt.de/.
Appendix C. Community repository for crawler extensions
In order to foster the re-usability of crawler definitions, we are building a community repository for crawler extensions, which can be found at https://gitlab.com/linkahead/crawler-extensions.
Appendix D. Software documentation
The official documentation for LinkAhead, including an installation guide, can be found at https://docs.indiscale.com. The documentation for the crawler framework that is presented in this article can be found at https://docs.indiscale.com/caosdb-crawler/.
Abbreviations, acronyms, and initialisms
- AI: artificial intelligence
- API: application programming interface
- DBMS: database management system
- ELN: electronic laboratory notebook
- ETL: extract, transform, and load
- FAIR: findable, accessible, interoperable, and reusable
- FDO: FAIR Digital Object
- GUI: graphical user interface
- IT: information technology
- ML: machine learning
- PID: persistent identifier
- RDBMS: relational database management system
- RDMS: research data management system
Acknowledgements
The authors thank Birte Hemmelskamp-Pfeiffer for providing information on https://dataportal.leibniz-zmt.de.
Author contributions
Conceptualization, A.S., H.t.W. and F.S.; software, A.S., H.t.W. and F.S.; resources, A.S., H.t.W., U.P., S.L. and F.S.; writing—original draft preparation, A.S., H.t.W. and F.S.; writing—review and editing, A.S., H.t.W., U.P., S.L. and F.S.; supervision, A.S. and H.t.W.; project administration, H.t.W. and S.L.; funding acquisition, A.S., U.P. and S.L. All authors have read and agreed to the published version of the manuscript.
Funding
A.S., U.P., and S.L. acknowledge financial support from the Volkswagen Stiftung within the framework of the project “Global Carbon Cycling and Complex Molecular Patterns in Aquatic Systems: Integrated Analyses Powered by Semantic Data Management”. S.L. acknowledges financial support from the DZHK and DFG SFB 1002 Modulary Units in Heart Failure, and the Else Kröner-Fresenius Foundation (EKFS).
Conflicts of interest
A.S. and H.t.W. are co-founders of IndiScale GmbH, a company providing commercial services for LinkAhead. Furthermore, H.t.W. and F.S. are employees of IndiScale GmbH. A.S. is external scientific advisor for IndiScale GmbH. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- ↑ 1.0 1.1 Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, IJsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship" (in en). Scientific Data 3 (1): 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. PMC PMC4792175. PMID 26978244. https://www.nature.com/articles/sdata201618.
- ↑ Deutsche Forschungsgemeinschaft (20 April 2022). "Guidelines for Safeguarding Good Research Practice. Code of Conduct" (in de). Zenodo. doi:10.5281/ZENODO.6472827. https://zenodo.org/record/6472827.
- ↑ Ferguson, Lea Maria; Bertelmann, Roland; Bruch, Christoph; Messerschmidt, Reinhard; Pampel, Heinz; Schrader, Antonia; Schultze-Motel, Paul; Weisweiler, Nina Leonie (2022) (in en). Good (Digital) Research Practice and Open Science Support and Best Practices for Implementing the DFG Code of Conduct “Guidelines for Safeguarding Good Research Practice”. Helmholtz Open Science Briefing. Good (Digital) Research Practice and Open Science Support and Best Practices for Implementing the DFG Code of Conduct “Guidelines for Safeguarding Good Research Practice” Version 2.0. pp. pages 19 pages470 kb. doi:10.34734/fzj-2023-03453. https://juser.fz-juelich.de/record/1014764.
- ↑ 4.0 4.1 4.2 Gray, Jim; Liu, David T.; Nieto-Santisteban, Maria; Szalay, Alex; DeWitt, David J.; Heber, Gerd (1 December 2005). "Scientific data management in the coming decade" (in en). ACM SIGMOD Record 34 (4): 34–41. doi:10.1145/1107499.1107503. ISSN 0163-5808. https://dl.acm.org/doi/10.1145/1107499.1107503.
- ↑ Samuel, Sheeba (2017), Blomqvist, Eva; Maynard, Diana; Gangemi, Aldo et al.., eds., "Integrative Data Management for Reproducibility of Microscopy Experiments" (in en), The Semantic Web (Cham: Springer International Publishing) 10250: 246–255, doi:10.1007/978-3-319-58451-5_19, ISBN 978-3-319-58450-8, https://link.springer.com/10.1007/978-3-319-58451-5_19. Retrieved 2024-06-11
- ↑ 6.0 6.1 6.2 Vaisman, Alejandro; Zimányi, Esteban (2014) (in en). Data Warehouse Systems: Design and Implementation. Berlin, Heidelberg: Springer Berlin Heidelberg. doi:10.1007/978-3-642-54655-6. ISBN 978-3-642-54654-9. https://link.springer.com/10.1007/978-3-642-54655-6.
- ↑ Barillari, Caterina; Ottoz, Diana S. M.; Fuentes-Serna, Juan Mariano; Ramakrishnan, Chandrasekhar; Rinn, Bernd; Rudolf, Fabian (15 February 2016). "openBIS ELN-LIMS: an open-source database for academic laboratories" (in en). Bioinformatics 32 (4): 638–640. doi:10.1093/bioinformatics/btv606. ISSN 1367-4811. PMC PMC4743625. PMID 26508761. https://academic.oup.com/bioinformatics/article/32/4/638/1743839.
- ↑ Hewera, Michael; Hänggi, Daniel; Gerlach, Björn; Kahlert, Ulf Dietrich (2 August 2021). "eLabFTW as an Open Science tool to improve the quality and translation of preclinical research" (in en). F1000Research 10: 292. doi:10.12688/f1000research.52157.3. ISSN 2046-1402. PMC PMC8323070. PMID 34381592. https://f1000research.com/articles/10-292/v3.
- ↑ Suhr, M.; Lehmann, C.; Bauer, C. R.; Bender, T.; Knopp, C.; Freckmann, L.; Öst Hansen, B.; Henke, C. et al. (1 December 2020). "Menoci: lightweight extensible web portal enhancing data management for biomedical research projects" (in en). BMC Bioinformatics 21 (1): 582. doi:10.1186/s12859-020-03928-1. ISSN 1471-2105. PMC PMC7745495. PMID 33334310. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03928-1.
- ↑ Bauch, Angela; Adamczyk, Izabela; Buczek, Piotr; Elmer, Franz-Josef; Enimanev, Kaloyan; Glyzewski, Pawel; Kohler, Manuel; Pylak, Tomasz et al. (1 December 2011). "openBIS: a flexible framework for managing and analyzing complex data in biology research" (in en). BMC Bioinformatics 12 (1): 468. doi:10.1186/1471-2105-12-468. ISSN 1471-2105. PMC PMC3275639. PMID 22151573. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-468.
- ↑ Dudchenko, Aleksei; Ringwald, Friedemann; Czernilofsky, Felix; Dietrich, Sascha; Knaup, Petra; Ganzinger, Matthias (25 May 2022), Séroussi, Brigitte; Weber, Patrick; Dhombres, Ferdinand et al.., eds., "Large-File Raw Data Synchronization for openBIS Research Repositories", Studies in Health Technology and Informatics (IOS Press), doi:10.3233/shti220486, ISBN 978-1-64368-284-6, https://ebooks.iospress.nl/doi/10.3233/SHTI220486. Retrieved 2024-06-11
- ↑ McBride, Brian (2004), Staab, Steffen; Studer, Rudi, eds., "The Resource Description Framework (RDF) and its Vocabulary Description Language RDFS" (in en), Handbook on Ontologies (Berlin, Heidelberg: Springer Berlin Heidelberg): 51–65, doi:10.1007/978-3-540-24750-0_3, ISBN 978-3-662-11957-0, http://link.springer.com/10.1007/978-3-540-24750-0_3. Retrieved 2024-06-11
- ↑ W3C OWL Working Group (11 December 2012). "OWL 2 Web Ontology Language Document Overview (Second Edition)". World Wide Web Consortium. https://www.w3.org/TR/owl2-overview/.
- ↑ Pérez, Jorge; Arenas, Marcelo; Gutierrez, Claudio (1 August 2009). "Semantics and complexity of SPARQL" (in en). ACM Transactions on Database Systems 34 (3): 1–45. doi:10.1145/1567274.1567278. ISSN 0362-5915. https://dl.acm.org/doi/10.1145/1567274.1567278.
- ↑ Bizer, C.; Heath, T.; Ayers, D. et al. (2007). "Interlinking Open Data on the Web" (PDF). Proceedings of the 4th European Semantic Web Conference. http://tomheath.com/papers/bizer-heath-eswc2007-interlinking-open-data.pdf.
- ↑ Bizer, Christian; Heath, Tom; Berners-Lee, Tim (1 July 2009). "Linked Data - The Story So Far:" (in ng). International Journal on Semantic Web and Information Systems 5 (3): 1–22. doi:10.4018/jswis.2009081901. ISSN 1552-6283. https://services.igi-global.com/resolvedoi/resolve.aspx?doi=10.4018/jswis.2009081901.
- ↑ 17.0 17.1 De Smedt, Koenraad; Koureas, Dimitris; Wittenburg, Peter (11 April 2020). "FAIR Digital Objects for Science: From Data Pieces to Actionable Knowledge Units" (in en). Publications 8 (2): 21. doi:10.3390/publications8020021. ISSN 2304-6775. https://www.mdpi.com/2304-6775/8/2/21.
- ↑ Vassiliadis, Panos (1 July 2009). "A Survey of Extract–Transform–Load Technology:" (in ng). International Journal of Data Warehousing and Mining 5 (3): 1–27. doi:10.4018/jdwm.2009070101. ISSN 1548-3924. https://services.igi-global.com/resolvedoi/resolve.aspx?doi=10.4018/jdwm.2009070101.
- ↑ 19.0 19.1 Fitschen, Timm; Schlemmer, Alexander; Hornung, Daniel; tom Wörden, Henrik; Parlitz, Ulrich; Luther, Stefan (10 June 2019). "CaosDB—Research Data Management for Complex, Changing, and Automated Research Workflows" (in en). Data 4 (2): 83. doi:10.3390/data4020083. ISSN 2306-5729. https://www.mdpi.com/2306-5729/4/2/83.
- ↑ Hornung, Daniel; Spreckelsen, Florian; Weiß, Thomas (30 January 2024) (in en). Agile Research Data Management with Open Source: LinkAhead. doi:10.48694/INGGRID.3866. ISSN 2941-1300. https://www.inggrid.org/article/id/3866/.
- ↑ 21.0 21.1 Spreckelsen, Florian; Rüchardt, Baltasar; Lebert, Jan; Luther, Stefan; Parlitz, Ulrich; Schlemmer, Alexander (24 April 2020). "Guidelines for a Standardized Filesystem Layout for Scientific Data" (in en). Data 5 (2): 43. doi:10.3390/data5020043. ISSN 2306-5729. https://www.mdpi.com/2306-5729/5/2/43.
- ↑ Gorgolewski, Krzysztof J.; Auer, Tibor; Calhoun, Vince D.; Craddock, R. Cameron; Das, Samir; Duff, Eugene P.; Flandin, Guillaume; Ghosh, Satrajit S. et al. (21 June 2016). "The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments" (in en). Scientific Data 3 (1): 160044. doi:10.1038/sdata.2016.44. ISSN 2052-4463. PMC PMC4978148. PMID 27326542. https://www.nature.com/articles/sdata201644.
- ↑ Mildenberger, Peter; Eichelberg, Marco; Martin, Eric (1 April 2002). "Introduction to the DICOM standard" (in en). European Radiology 12 (4): 920–927. doi:10.1007/s003300101100. ISSN 0938-7994. http://link.springer.com/10.1007/s003300101100.
- ↑ 24.0 24.1 Koranne, Sandeep (2011), "Hierarchical Data Format 5 : HDF5" (in en), Handbook of Open Source Tools (Boston, MA: Springer US): 191–200, doi:10.1007/978-1-4419-7719-9_10, ISBN 978-1-4419-7718-2, https://link.springer.com/10.1007/978-1-4419-7719-9_10. Retrieved 2024-06-11
- ↑ 25.0 25.1 Folk, Mike; Heber, Gerd; Koziol, Quincey; Pourmal, Elena; Robinson, Dana (25 March 2011). "An overview of the HDF5 technology suite and its applications" (in en). Proceedings of the EDBT/ICDT 2011 Workshop on Array Databases (Uppsala Sweden: ACM): 36–47. doi:10.1145/1966895.1966900. ISBN 978-1-4503-0614-0. https://dl.acm.org/doi/10.1145/1966895.1966900.
- ↑ Schlemmer, Alexander (5 May 2021), "Mapping data files to semantic data models using the CaosDB crawler" (in en), Zenodo, doi:10.5281/zenodo.8246645, https://zenodo.org/record/8246645. Retrieved 2024-06-11
- ↑ Pezoa, Felipe; Reutter, Juan L.; Suarez, Fernando; Ugarte, Martín; Vrgoč, Domagoj (11 April 2016). "Foundations of JSON Schema" (in en). Proceedings of the 25th International Conference on World Wide Web (Montréal Québec Canada: International World Wide Web Conferences Steering Committee): 263–273. doi:10.1145/2872427.2883029. ISBN 978-1-4503-4143-1. https://dl.acm.org/doi/10.1145/2872427.2883029.
- ↑ Internet Engineering Task Force (IETF) (21 January 2020). "The JavaScript Object Notation (JSON) Data Interchange Format - RFC 7159". Datatracker. https://datatracker.ietf.org/doc/rfc7159/. Retrieved 10 August 2023.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.