Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Asiimwe JofTransMed21 19.png|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:From biobank and data silos into a data commons: Convergence to support translational medicine|From biobank and data silos into a data commons: Convergence to support translational medicine]]"''' | ||
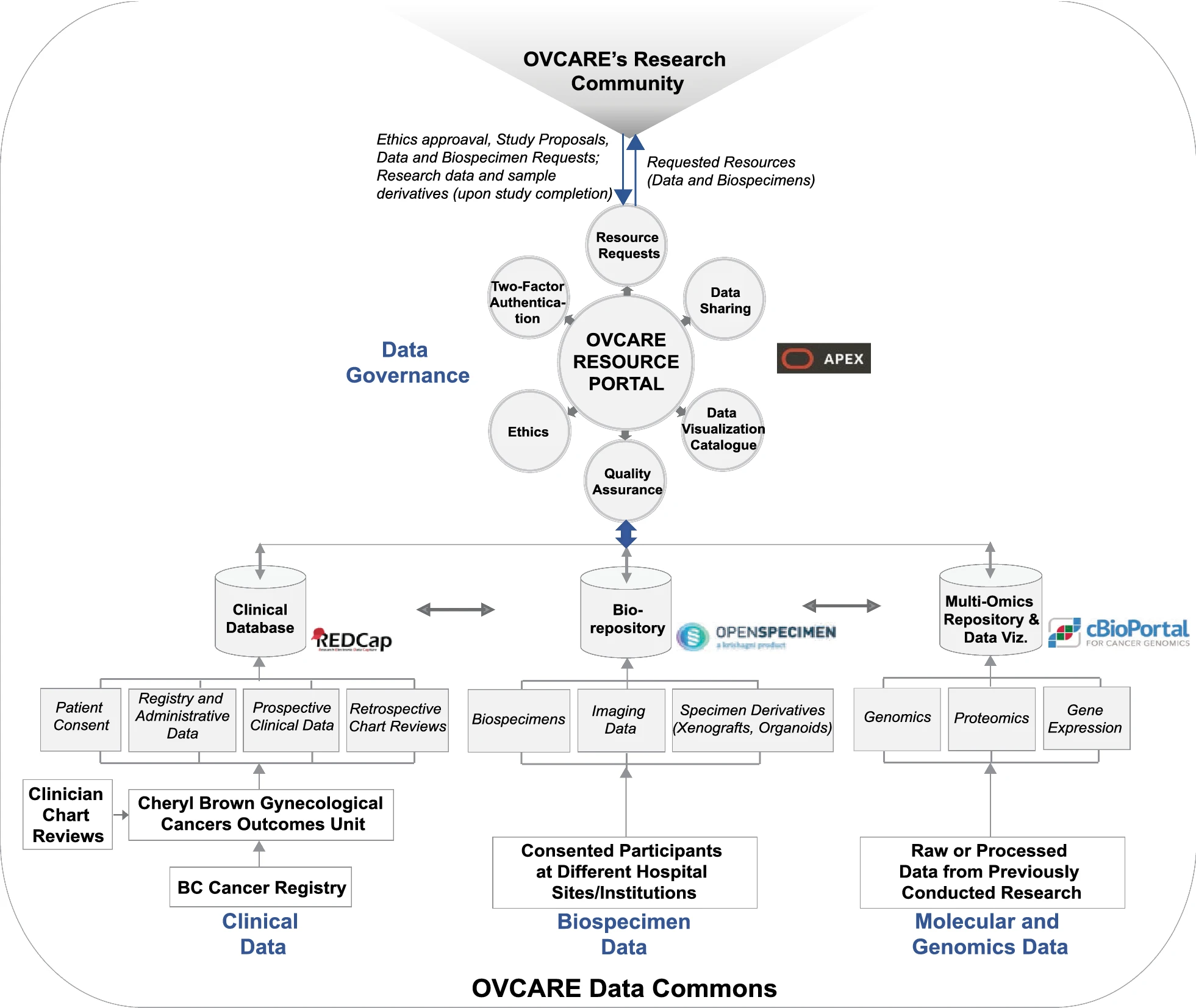
To drive [[Translational research|translational medicine]], modern day [[biobank]]s need to integrate with other sources of data (e.g., [[Health informatics|clinical]], [[genomics]]) to support novel data-intensive research. Currently, vast amounts of research and clinical data remain in silos, held and managed by individual researchers, operating under different standards and governance structures; such a framework impedes sharing and effective use of data. In this article, we describe the journey of British Columbia’s Gynecological Cancer Research Program (OVCARE) in moving a traditional tumor biobank, outcomes unit, and a collection of data silos into an integrated [[Open data#Policies and strategies|data commons]] to support data standardization and [[Data sharing|resource sharing]] under collaborative governance, as a means of providing the gynecologic cancer research community in British Columbia access to tissue samples and associated clinical and [[Molecular diagnostics|molecular]] data from thousands of patients ... ('''[[Journal:From biobank and data silos into a data commons: Convergence to support translational medicine|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
{{flowlist | | {{flowlist | | ||
* [[Journal:Quality management system implementation in human and animal laboratories|Quality management system implementation in human and animal laboratories]] | |||
* [[Journal:Data management and modeling in plant biology|Data management and modeling in plant biology]] | * [[Journal:Data management and modeling in plant biology|Data management and modeling in plant biology]] | ||
* [[Journal:Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences|Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences]] | * [[Journal:Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences|Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences]] | ||
}} | }} | ||
Revision as of 14:07, 22 August 2022
"From biobank and data silos into a data commons: Convergence to support translational medicine"
To drive translational medicine, modern day biobanks need to integrate with other sources of data (e.g., clinical, genomics) to support novel data-intensive research. Currently, vast amounts of research and clinical data remain in silos, held and managed by individual researchers, operating under different standards and governance structures; such a framework impedes sharing and effective use of data. In this article, we describe the journey of British Columbia’s Gynecological Cancer Research Program (OVCARE) in moving a traditional tumor biobank, outcomes unit, and a collection of data silos into an integrated data commons to support data standardization and resource sharing under collaborative governance, as a means of providing the gynecologic cancer research community in British Columbia access to tissue samples and associated clinical and molecular data from thousands of patients ... (Full article...)
Recently featured: