Difference between revisions of "Main Page/Featured article of the week/2017"
Shawndouglas (talk | contribs) (Added last week's article of the week.) |
Shawndouglas (talk | contribs) (Added last week's article of the week.) |
||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 23–29:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 30–February 05:</h2> | ||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Tsai JofPersMed2016 6-1.png|240px]]</div> | |||
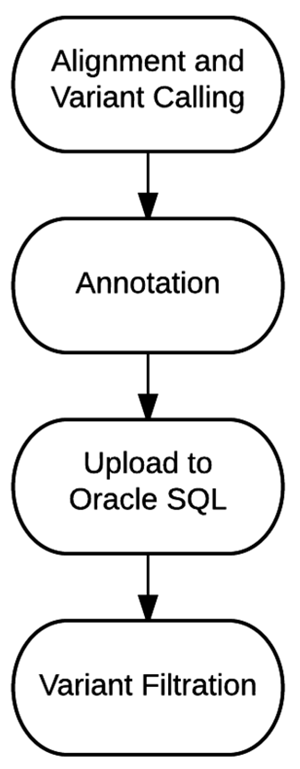
'''"[[Journal:Bioinformatics workflow for clinical whole genome sequencing at Partners HealthCare Personalized Medicine|Bioinformatics workflow for clinical whole genome sequencing at Partners HealthCare Personalized Medicine]]"''' | |||
Effective implementation of precision medicine will be enhanced by a thorough understanding of each patient’s genetic composition to better treat his or her presenting symptoms or mitigate the onset of disease. This ideally includes the sequence [[information]] of a complete genome for each individual. At Partners HealthCare Personalized Medicine, we have developed a clinical process for whole genome sequencing (WGS) with application in both healthy individuals and those with disease. In this manuscript, we will describe our [[bioinformatics]] strategy to efficiently process and deliver [[Genomics|genomic]] data to geneticists for clinical interpretation. We describe the handling of data from FASTQ to the final variant list for clinical review for the final report. We will also discuss our methodology for validating this workflow and the cost implications of running WGS. ('''[[Journal:Bioinformatics workflow for clinical whole genome sequencing at Partners HealthCare Personalized Medicine|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 23–29:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | <div style="padding:0.4em 1em 0.3em 1em;"> | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig5 Ye JofPathInformatics2016 7.jpg|240px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig5 Ye JofPathInformatics2016 7.jpg|240px]]</div> | ||
Revision as of 16:04, 7 February 2017
|
|
If you're looking for other "Article of the Week" archives: 2014 - 2015 - 2016 - 2017 |
Featured article of the week archive - 2017
Welcome to the LIMSwiki 2017 archive for the Featured Article of the Week.
Featured article of the week: January 30–February 05:Effective implementation of precision medicine will be enhanced by a thorough understanding of each patient’s genetic composition to better treat his or her presenting symptoms or mitigate the onset of disease. This ideally includes the sequence information of a complete genome for each individual. At Partners HealthCare Personalized Medicine, we have developed a clinical process for whole genome sequencing (WGS) with application in both healthy individuals and those with disease. In this manuscript, we will describe our bioinformatics strategy to efficiently process and deliver genomic data to geneticists for clinical interpretation. We describe the handling of data from FASTQ to the final variant list for clinical review for the final report. We will also discuss our methodology for validating this workflow and the cost implications of running WGS. (Full article...)
|