Difference between revisions of "File:Fig1 Goldberg mBio2015 6-6.jpg"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Added summary.) |
||
| Line 1: | Line 1: | ||
==Summary== | |||
{{Information | |||
|Description='''Figure 1.''' NGS genome analysis. The general process of using NGS for analysis of a single genome is depicted in this figure. Note that there are many variations on this approach. Purified DNA (i.e., input genome) is fragmented and run through the DNA sequencing process. The sequencing instrument produces either short (e.g., for Illumina) or long (e.g., for Pacific Biosciences) sequence reads depending on the platform used. When the goal is to produce a complete genome (e.g., for identifying virulence or antibiotic resistance genes or for comparative genomic studies), the reads are assembled into genomes using specialized software. Some gaps in the assembly may occur, leading to a draft genome sequence composed of many contigs. When the genome is assembled without gaps, it is said to be closed. When the goal is to identify variants (e.g., SNPs) with respect to a reference genome, the reads can be aligned directly with the reference genome and sequence variants can be identified using specialized programs. These variants serve to define the organism at the subspecies and strain level and are useful in epidemiological tracking as well as to identify mutations that occur. | |||
|Source={{cite journal |title=Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics |journal=mBio |author=Goldberg, Brittany; Sichtig, Heike; Geyer, Chelsie; Ledeboer, Nathan; Weinstock, George M. |volume=6 |issue=6 |pages=e01888-15 |year=2015 |doi=10.1128/mBio.01888-15 |pmid=26646014 |pmc=PMC4669390}} | |||
|Author=Goldberg, Brittany; Sichtig, Heike; Geyer, Chelsie; Ledeboer, Nathan; Weinstock, George M. | |||
|Date=2016 | |||
|Permission=[http://creativecommons.org/licenses/by-nc-sa/3.0/ Creative Commons Attribution-Noncommercial-ShareAlike 3.0 Unported] | |||
}} | |||
== Licensing == | == Licensing == | ||
{{cc-by-nc-sa-3.0}} | {{cc-by-nc-sa-3.0}} | ||
Latest revision as of 16:29, 20 September 2016
Summary
| Description |
Figure 1. NGS genome analysis. The general process of using NGS for analysis of a single genome is depicted in this figure. Note that there are many variations on this approach. Purified DNA (i.e., input genome) is fragmented and run through the DNA sequencing process. The sequencing instrument produces either short (e.g., for Illumina) or long (e.g., for Pacific Biosciences) sequence reads depending on the platform used. When the goal is to produce a complete genome (e.g., for identifying virulence or antibiotic resistance genes or for comparative genomic studies), the reads are assembled into genomes using specialized software. Some gaps in the assembly may occur, leading to a draft genome sequence composed of many contigs. When the genome is assembled without gaps, it is said to be closed. When the goal is to identify variants (e.g., SNPs) with respect to a reference genome, the reads can be aligned directly with the reference genome and sequence variants can be identified using specialized programs. These variants serve to define the organism at the subspecies and strain level and are useful in epidemiological tracking as well as to identify mutations that occur. |
|---|---|
| Source |
Goldberg, Brittany; Sichtig, Heike; Geyer, Chelsie; Ledeboer, Nathan; Weinstock, George M. (2015). "Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics". mBio 6 (6): e01888-15. doi:10.1128/mBio.01888-15. PMC PMC4669390. PMID 26646014. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4669390. |
| Date |
2016 |
| Author |
Goldberg, Brittany; Sichtig, Heike; Geyer, Chelsie; Ledeboer, Nathan; Weinstock, George M. |
| Permission (Reusing this file) |
Creative Commons Attribution-Noncommercial-ShareAlike 3.0 Unported |
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 16:25, 20 September 2016 | 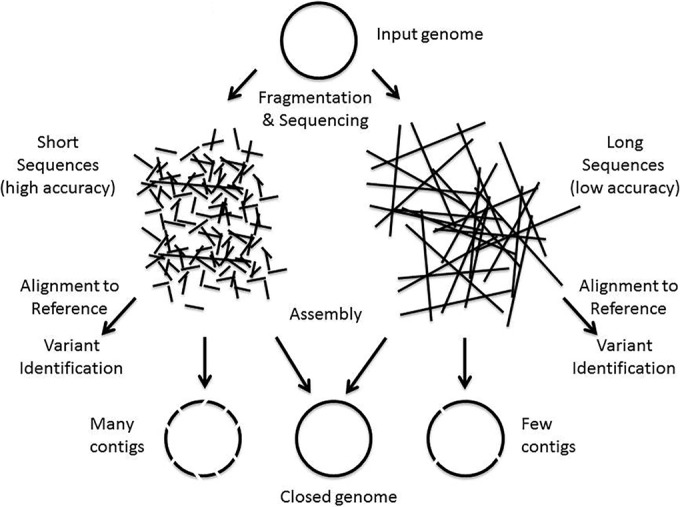 | 680 × 507 (63 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.









