Difference between revisions of "Main Page/Featured article of the week/2016"
Shawndouglas (talk | contribs) (Added last week's article of the week.) |
Shawndouglas (talk | contribs) (Added last week's article of the week.) |
||
| Line 17: | Line 17: | ||
<!-- Below this line begin pasting previous news --> | <!-- Below this line begin pasting previous news --> | ||
<h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 15–21:</h2> | <h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 22–28:</h2> | ||
<div style="padding:0.4em 1em 0.3em 1em;"> | |||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 OConnor BMCInformatics2010 11-12.jpg|220px]]</div> | |||
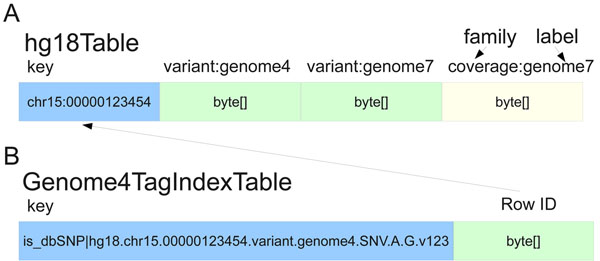
'''"[[Journal:SeqWare Query Engine: Storing and searching sequence data in the cloud|SeqWare Query Engine: Storing and searching sequence data in the cloud]]"''' | |||
Since the introduction of next-generation DNA sequencers the rapid increase in [[Sequencing|sequencer]] throughput, and associated drop in costs, has resulted in more than a dozen human [[Genomics|genomes]] being resequenced over the last few years. These efforts are merely a prelude for a future in which genome resequencing will be commonplace for both biomedical research and clinical applications. The dramatic increase in sequencer output strains all facets of computational infrastructure, especially databases and query interfaces. The advent of cloud computing, and a variety of powerful tools designed to process petascale datasets, provide a compelling solution to these ever increasing demands. | |||
In this work, we present the [[SeqWare]] Query Engine which has been created using modern cloud computing technologies and designed to support databasing information from thousands of genomes. Our backend implementation was built using the highly scalable, NoSQL HBase database from the Hadoop project. We also created a web-based frontend that provides both a programmatic and interactive query interface and integrates with widely used genome browsers and tools. ('''[[Journal:SeqWare Query Engine: Storing and searching sequence data in the cloud|Full article...]]''')<br /> | |||
</div> | |||
|- | |||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: February 15–21:</h2> | |||
<div style="padding:0.4em 1em 0.3em 1em;"> | <div style="padding:0.4em 1em 0.3em 1em;"> | ||
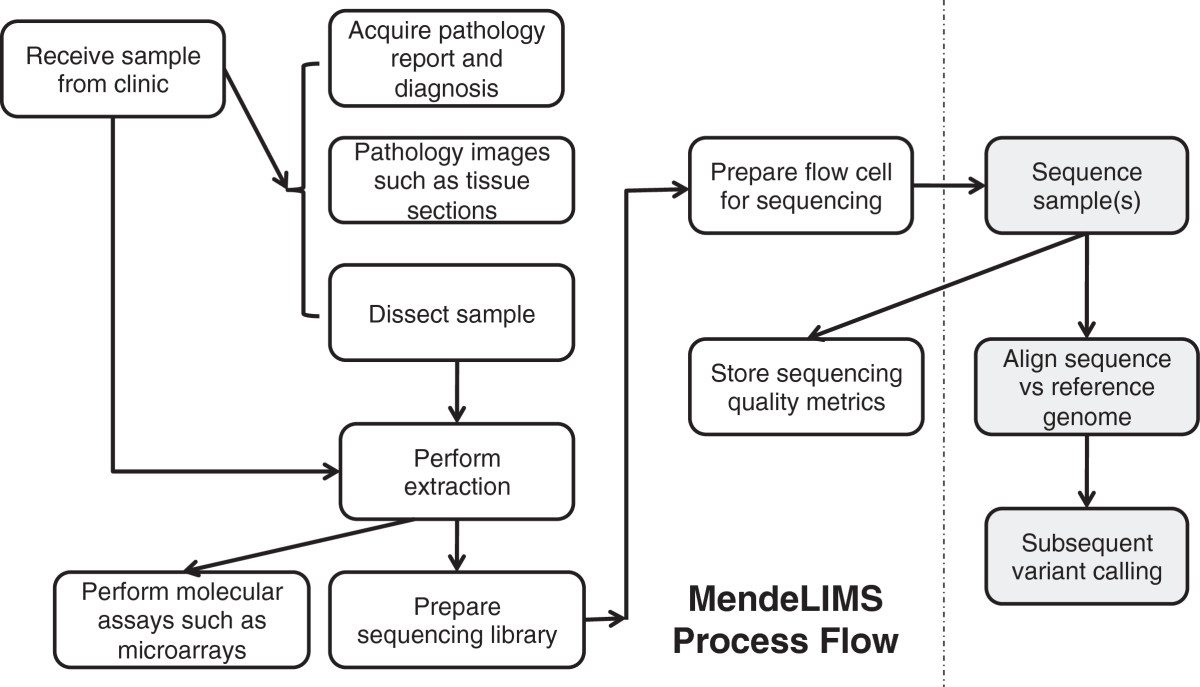
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 SinghBMCBioinformatics2015 12-6.png|220px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 SinghBMCBioinformatics2015 12-6.png|220px]]</div> | ||
Revision as of 16:14, 29 February 2016
|
|
If you're looking for the 2014 archive, it can be found here. The 2015 archive is here. |
Featured article of the week archive - 2016
Welcome to the LIMSwiki 2016 archive for the Featured Article of the Week.
Featured article of the week: February 22–28:"SeqWare Query Engine: Storing and searching sequence data in the cloud" Since the introduction of next-generation DNA sequencers the rapid increase in sequencer throughput, and associated drop in costs, has resulted in more than a dozen human genomes being resequenced over the last few years. These efforts are merely a prelude for a future in which genome resequencing will be commonplace for both biomedical research and clinical applications. The dramatic increase in sequencer output strains all facets of computational infrastructure, especially databases and query interfaces. The advent of cloud computing, and a variety of powerful tools designed to process petascale datasets, provide a compelling solution to these ever increasing demands. In this work, we present the SeqWare Query Engine which has been created using modern cloud computing technologies and designed to support databasing information from thousands of genomes. Our backend implementation was built using the highly scalable, NoSQL HBase database from the Hadoop project. We also created a web-based frontend that provides both a programmatic and interactive query interface and integrates with widely used genome browsers and tools. (Full article...)
|