Difference between revisions of "Journal:Digitalization concepts in academic bioprocess development"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Finished adding rest of content) |
||
| Line 18: | Line 18: | ||
|website = [https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/elsc.202300238 https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/elsc.202300238] | |website = [https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/elsc.202300238 https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/elsc.202300238] | ||
|download = [https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/pdfdirect/10.1002/elsc.202300238?download=true https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/pdfdirect/10.1002/elsc.202300238] (PDF) | |download = [https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/pdfdirect/10.1002/elsc.202300238?download=true https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/pdfdirect/10.1002/elsc.202300238] (PDF) | ||
}} | }} | ||
==Abstract== | ==Abstract== | ||
| Line 31: | Line 25: | ||
==Introduction== | ==Introduction== | ||
In contrast to industry, academic [[research]] [[Laboratory|laboratories]] require more flexibility than production lines. Besides the need for flexibility, loss of knowledge due to high staff turnover in universities is another challenge that makes full laboratory digitalization hard to achieve. | In contrast to industry, academic [[research]] [[Laboratory|laboratories]] require more flexibility than production lines. Besides the need for flexibility, loss of knowledge due to high staff turnover in universities is another challenge that makes full laboratory digitalization hard to achieve.<ref name=":0">{{Cite journal |last=Hollricher, K. |year=2022 |title=Digitales Lab - Viel luft nach oben |url=https://sila-standard.com/wp-content/uploads/2022/11/Digitales-Lab-viel-Luft-nach-oben.pdf |journal=Carl |volume=1 |pages=42–5}}</ref><ref name=":1">{{Cite journal |last=Neubauer |first=Peter |last2=Glauche |first2=Florian |last3=Cruz‐Bournazou |first3=M. Nicolas |date=2017-11 |title=Editorial: Bioprocess Development in the era of digitalization |url=https://onlinelibrary.wiley.com/doi/10.1002/elsc.201770113 |journal=Engineering in Life Sciences |language=en |volume=17 |issue=11 |pages=1140–1141 |doi=10.1002/elsc.201770113 |issn=1618-0240 |pmc=PMC6999283 |pmid=32624741}}</ref><ref>{{Citation |last=Jochums |first=Max |last2=Reinders |first2=Lars M. H. |last3=Tuerk |first3=Jochen |last4=Teutenberg |first4=Thorsten |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=Flexible Digitization of Highly Individualized Workflows Demonstrated Through the Quality Control of Patient-Specific Cytostatic Application Bags: Digitization from the Perspective of Small and Medium-Sized Laboratories |url=https://link.springer.com/10.1007/10_2021_190 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=115–129 |doi=10.1007/10_2021_190 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref><ref>{{Cite journal |last=Lütjohann |first=Dominic S. |last2=Jung |first2=Nicole |last3=Bräse |first3=Stefan |date=2015-05 |title=Open source life science automation: Design of experiments and data acquisition via “dial-a-device” |url=https://linkinghub.elsevier.com/retrieve/pii/S0169743915000842 |journal=Chemometrics and Intelligent Laboratory Systems |language=en |volume=144 |pages=100–107 |doi=10.1016/j.chemolab.2015.04.002}}</ref> "Digitalization" (what we'll be addressing) refers to an entire process, [[workflow]], or laboratory infrastructure, whereas "digitization" refers only to the procedure of converting something analog to a digital format (e.g., digitizing a standard operating procedure [SOP] from a piece of paper to a digital file).<ref name=":2">{{Cite journal |last=Kockmann |first=Norbert |date=2019 |title=Digital methods and tools for chemical equipment and plants |url=https://xlink.rsc.org/?DOI=C9RE00017H |journal=Reaction Chemistry & Engineering |language=en |volume=4 |issue=9 |pages=1522–1529 |doi=10.1039/C9RE00017H |issn=2058-9883}}</ref><ref>{{Cite journal |last=Krüger |first=Anna |last2=Schäfers |first2=Christian |last3=Busch |first3=Philip |last4=Antranikian |first4=Garabed |date=2020-11 |title=Digitalization in microbiology – Paving the path to sustainable circular bioeconomy |url=https://linkinghub.elsevier.com/retrieve/pii/S1871678420301497 |journal=New Biotechnology |language=en |volume=59 |pages=88–96 |doi=10.1016/j.nbt.2020.06.004}}</ref> Working in digital laboratories has the potential for error reduction, prevention of data loss, improved [[data integrity]], faster workflow development times, possible reduction of chemical and material use, and higher [[Sample (material)|sample]] throughput, leading to modern, transparent, and reproducible research and biomanufacturing.<ref name=":3">{{Cite journal |last=Richter |first=Jannik |last2=Lange |first2=Ferdinand |last3=Scheper |first3=Thomas |last4=Solle |first4=Dörte |last5=Beutel |first5=Sascha |date=2023-04 |title=Digitale Zwillinge in der Bioprozesstechnik – Chancen und Möglichkeiten |url=https://onlinelibrary.wiley.com/doi/10.1002/cite.202200166 |journal=Chemie Ingenieur Technik |language=en |volume=95 |issue=4 |pages=498–510 |doi=10.1002/cite.202200166 |issn=0009-286X}}</ref><ref name=":4">{{Citation |last=Thurow |first=Kerstin |date=2021 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=Automation for Life Science Laboratories |url=https://link.springer.com/10.1007/10_2021_170 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=3–22 |doi=10.1007/10_2021_170 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref><ref name=":5">{{Cite journal |last=Schmid |first=Ingrid |last2=Aschoff |first2=Joachim |date=2017-11 |title=A scalable software framework for data integration in bioprocess development |url=https://onlinelibrary.wiley.com/doi/10.1002/elsc.201600008 |journal=Engineering in Life Sciences |language=en |volume=17 |issue=11 |pages=1159–1165 |doi=10.1002/elsc.201600008 |issn=1618-0240 |pmc=PMC6999508 |pmid=32624743}}</ref><ref>{{Cite journal |last=Neubauer |first=Peter |last2=Cruz |first2=Nicolas |last3=Glauche |first3=Florian |last4=Junne |first4=Stefan |last5=Knepper |first5=Andreas |last6=Raven |first6=Michael |date=2013-05 |title=Consistent development of bioprocesses from microliter cultures to the industrial scale |url=https://onlinelibrary.wiley.com/doi/10.1002/elsc.201200021 |journal=Engineering in Life Sciences |language=en |volume=13 |issue=3 |pages=224–238 |doi=10.1002/elsc.201200021 |issn=1618-0240}}</ref><ref name=":6">{{Cite journal |last=Narayanan |first=Harini |last2=Luna |first2=Martin F. |last3=von Stosch |first3=Moritz |last4=Cruz Bournazou |first4=Mariano Nicolas |last5=Polotti |first5=Gianmarco |last6=Morbidelli |first6=Massimo |last7=Butté |first7=Alessandro |last8=Sokolov |first8=Michael |date=2020-01 |title=Bioprocessing in the Digital Age: The Role of Process Models |url=https://onlinelibrary.wiley.com/doi/10.1002/biot.201900172 |journal=Biotechnology Journal |language=en |volume=15 |issue=1 |pages=1900172 |doi=10.1002/biot.201900172 |issn=1860-6768}}</ref><ref name=":7">{{Cite journal |last=Saha |first=Himadri Nath |last2=Auddy |first2=Supratim |last3=Pal |first3=Subrata |last4=Kumar |first4=Shubham |last5=Jasu |first5=Subhadeep |last6=Singh |first6=Rocky |last7=Singh |first7=Rakhee |last8=Banerjee |first8=Swarnadeep |last9=Sharan |first9=Priyanshu |last10=Maity |first10=Ankita |date=2017-08 |title=Internet of Things (IoT) on bio-technology |url=http://ieeexplore.ieee.org/document/8079624/ |journal=2017 8th Annual Industrial Automation and Electromechanical Engineering Conference (IEMECON) |publisher=IEEE |place=Bangkok, Thailand |pages=364–369 |doi=10.1109/IEMECON.2017.8079624 |isbn=978-1-5386-2215-5}}</ref><ref>{{Cite journal |last=Osthege |first=Michael |last2=Tenhaef |first2=Niklas |last3=Zyla |first3=Rebecca |last4=Müller |first4=Carolin |last5=Hemmerich |first5=Johannes |last6=Wiechert |first6=Wolfgang |last7=Noack |first7=Stephan |last8=Oldiges |first8=Marco |date=2022-03 |title=bletl ‐ A Python package for integrating BioLector microcultivation devices in the Design‐Build‐Test‐Learn cycle |url=https://onlinelibrary.wiley.com/doi/10.1002/elsc.202100108 |journal=Engineering in Life Sciences |language=en |volume=22 |issue=3-4 |pages=242–259 |doi=10.1002/elsc.202100108 |issn=1618-0240 |pmc=PMC8961055 |pmid=35382539}}</ref><ref>{{Cite journal |last=Solle |first=Dörte |last2=Hitzmann |first2=Bernd |last3=Herwig |first3=Christoph |last4=Pereira Remelhe |first4=Manuel |last5=Ulonska |first5=Sophia |last6=Wuerth |first6=Lynn |last7=Prata |first7=Adrian |last8=Steckenreiter |first8=Thomas |date=2017-05 |title=Between the Poles of Data‐Driven and Mechanistic Modeling for Process Operation |url=https://onlinelibrary.wiley.com/doi/10.1002/cite.201600175 |journal=Chemie Ingenieur Technik |language=en |volume=89 |issue=5 |pages=542–561 |doi=10.1002/cite.201600175 |issn=0009-286X}}</ref> | ||
Developing a digitalization strategy for an academic [[bioprocess]] laboratory is an interdisciplinary task. Laboratory workers have to work closely with IT experts in order to achieve a digital infrastructure that can be maintained and is supporting their laboratory work rather than hindering it. | Developing a digitalization strategy for an academic [[bioprocess]] laboratory is an interdisciplinary task. Laboratory workers have to work closely with IT experts in order to achieve a digital infrastructure that can be maintained and is supporting their laboratory work rather than hindering it.<ref name=":2" /><ref name=":3" /><ref name=":5" /><ref>{{Cite journal |last=Wong |first=Sybil |last2=Pan |first2=Mingke |last3=Shaw |first3=Allen |last4=Gershater |first4=Markus |date=2022-07 |title=What digitalization in biology R&D means for biotech companies and life scientists |url=https://www.nature.com/articles/s41587-022-01309-y |journal=Nature Biotechnology |language=en |volume=40 |issue=7 |pages=1151–1153 |doi=10.1038/s41587-022-01309-y |issn=1087-0156}}</ref><ref>{{Cite journal |last=Weiss |first=Charles J. |date=2017-05-09 |title=Scientific Computing for Chemists: An Undergraduate Course in Simulations, Data Processing, and Visualization |url=https://pubs.acs.org/doi/10.1021/acs.jchemed.7b00078 |journal=Journal of Chemical Education |language=en |volume=94 |issue=5 |pages=592–597 |doi=10.1021/acs.jchemed.7b00078 |issn=0021-9584}}</ref> This kind of collaboration is rarely found in academic bioprocess laboratories, so even the smallest digitalization task can be a major challenge. Hardware vendors selling their devices with proprietary software and restrictive access and [[information]] about their [[Interface (computing)|interfaces]] is making digitalization even more difficult.<ref name=":0" /><ref name=":3" /><ref name=":8">{{Cite web |last=Kraus, V.P. |date=11 October 2021 |title=Von OPC UA zu LADS: Ein Kommunikationsstandard entsteht |work=Labor Praxis |url=https://www.laborpraxis.vogel.de/von-opc-ua-zu-lads-ein-kommunikationsstandard-entsteht-a-13c637c18db4e91c678b4734c0c6b941/ |publisher=Vogel Communications Group}}</ref> Therefore, academic researchers working in the field of digitalization are developing concepts that are fitting their individual needs at the time. This is why there are manifold digitalization concepts throughout academic laboratories. | ||
This review will present different digitalization concepts throughout academic bioprocess development laboratories. Before looking at different individual concepts in more detail, general aspects like [[Journal:The FAIR Guiding Principles for scientific data management and stewardship|FAIR data]] (data that is readily findable, accessible, interoperable, and reusable), standard communication protocols, [[digital twin]]s, and interaction with laboratory devices will be addressed. | This review will present different digitalization concepts throughout academic bioprocess development laboratories. Before looking at different individual concepts in more detail, general aspects like [[Journal:The FAIR Guiding Principles for scientific data management and stewardship|FAIR data]] (data that is readily findable, accessible, interoperable, and reusable), standard communication protocols, [[digital twin]]s, and interaction with laboratory devices will be addressed. | ||
| Line 39: | Line 33: | ||
==Basic concepts of digitalization== | ==Basic concepts of digitalization== | ||
===FAIR data=== | ===FAIR data=== | ||
With the progressing digitalization of laboratories, the generated amount of data is steadily increasing. Therefore, good [[Information management|data management]] systems will become inevitable in bioprocess development laboratories. The first step towards good data management is storing the data and [[metadata]] according to the FAIR data principles. | With the progressing digitalization of laboratories, the generated amount of data is steadily increasing. Therefore, good [[Information management|data management]] systems will become inevitable in bioprocess development laboratories. The first step towards good data management is storing the data and [[metadata]] according to the FAIR data principles.<ref name=":9">{{Cite book |last=European Commission. Directorate General for Research and Innovation. |date=2018 |title=Turning FAIR into reality: final report and action plan from the European Commission expert group on FAIR data. |url=https://data.europa.eu/doi/10.2777/1524 |publisher=Publications Office |place=LU |doi=10.2777/1524}}</ref> Both humans and machines should be able to find the data with metadata and a clear unique identifier. The data should be digitally accessible for the user with the appropriate tool. It should be noted that accessible in this context does not mean that the data are "open" or "free" but that transparency around the used data concerning its availability is given.<ref>{{Cite journal |last=Landi |first=Annalisa |last2=Thompson |first2=Mark |last3=Giannuzzi |first3=Viviana |last4=Bonifazi |first4=Fedele |last5=Labastida |first5=Ignasi |last6=da Silva Santos |first6=Luiz Olavo Bonino |last7=Roos |first7=Marco |date=2020-01 |title=The “A” of FAIR – As Open as Possible, as Closed as Necessary |url=https://direct.mit.edu/dint/article/2/1-2/47-55/9998 |journal=Data Intelligence |language=en |volume=2 |issue=1-2 |pages=47–55 |doi=10.1162/dint_a_00027 |issn=2641-435X}}</ref><ref>{{Cite journal |last=Mons |first=Barend |last2=Neylon |first2=Cameron |last3=Velterop |first3=Jan |last4=Dumontier |first4=Michel |last5=da Silva Santos |first5=Luiz Olavo Bonino |last6=Wilkinson |first6=Mark D. |date=2017-03-07 |title=Cloudy, increasingly FAIR; revisiting the FAIR Data guiding principles for the European Open Science Cloud |url=https://www.medra.org/servlet/aliasResolver?alias=iospress&doi=10.3233/ISU-170824 |journal=Information Services & Use |volume=37 |issue=1 |pages=49–56 |doi=10.3233/ISU-170824}}</ref> Interoperable data means data are presented using vocabulary and data encoding that follows the FAIR data principles and can be read by machines. In order for data to be reusable they need to be rich in metadata and descriptive documentation on the circumstances in which the data were generated. Rich metadata should ideally describe the data in a meaningful way, including the setup and context of the experiment, technical setting, and information about the provenance.<ref name=":4" /><ref name=":9" /><ref name=":10">{{Cite journal |last=Solle |first=Dörte |date=2020-07 |title=Be FAIR to your data |url=https://link.springer.com/10.1007/s00216-020-02526-7 |journal=Analytical and Bioanalytical Chemistry |language=en |volume=412 |issue=17 |pages=3961–3965 |doi=10.1007/s00216-020-02526-7 |issn=1618-2642 |pmc=PMC7320032 |pmid=32300841}}</ref><ref>{{Cite journal |last=Wilkinson |first=Mark D. |last2=Dumontier |first2=Michel |last3=Aalbersberg |first3=IJsbrand Jan |last4=Appleton |first4=Gabrielle |last5=Axton |first5=Myles |last6=Baak |first6=Arie |last7=Blomberg |first7=Niklas |last8=Boiten |first8=Jan-Willem |last9=da Silva Santos |first9=Luiz Bonino |last10=Bourne |first10=Philip E. |last11=Bouwman |first11=Jildau |date=2016-03-15 |title=The FAIR Guiding Principles for scientific data management and stewardship |url=https://www.nature.com/articles/sdata201618 |journal=Scientific Data |language=en |volume=3 |issue=1 |pages=160018 |doi=10.1038/sdata.2016.18 |issn=2052-4463 |pmc=PMC4792175 |pmid=26978244}}</ref><ref>{{Cite journal |last=Atici |first=Levent |last2=Kansa |first2=Sarah Whitcher |last3=Lev-Tov |first3=Justin |last4=Kansa |first4=Eric C. |date=2013-12 |title=Other People’s Data: A Demonstration of the Imperative of Publishing Primary Data |url=http://link.springer.com/10.1007/s10816-012-9132-9 |journal=Journal of Archaeological Method and Theory |language=en |volume=20 |issue=4 |pages=663–681 |doi=10.1007/s10816-012-9132-9 |issn=1072-5369}}</ref> | ||
Most research data are currently stored locally and not organized in a standardized way. Storing data according to FAIR data principles allows them to be read by machines and not just the human laboratory worker who generated them. Other advantages of following the FAIR data principles, as well as publishing research data, is that this data will stay available over time. Available or even openly online shared data, including metadata, can help to determine if the publication is of high quality. It can be reproduced, reanalyzed, used for new analyses, or even compared to or combined with other data promoting a deeper understanding of the topic or perhaps generating new knowledge. | Most research data are currently stored locally and not organized in a standardized way. Storing data according to FAIR data principles allows them to be read by machines and not just the human laboratory worker who generated them. Other advantages of following the FAIR data principles, as well as publishing research data, is that this data will stay available over time. Available or even openly online shared data, including metadata, can help to determine if the publication is of high quality. It can be reproduced, reanalyzed, used for new analyses, or even compared to or combined with other data promoting a deeper understanding of the topic or perhaps generating new knowledge.<ref name=":7" /><ref name=":10" /><ref>{{Citation |last=Neumann |first=Janna |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=FAIR Data Infrastructure |url=https://link.springer.com/10.1007/10_2021_193 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=195–207 |doi=10.1007/10_2021_193 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref> | ||
Before computers were used on a large scale in laboratories, results of experiments were handwritten into [[Laboratory notebook|paper notebooks]]. This is still happening in many cases in today's bioprocess development laboratories, although the use of [[electronic laboratory notebook]]s (ELNs) is emerging quickly. | Before computers were used on a large scale in laboratories, results of experiments were handwritten into [[Laboratory notebook|paper notebooks]]. This is still happening in many cases in today's bioprocess development laboratories, although the use of [[electronic laboratory notebook]]s (ELNs) is emerging quickly.<ref name=":11">{{Cite journal |last=Dunie |first=Matt |date=2017-11-07 |editor-last=Lawlor |editor-first=Bonnie |title=The importance of research data management: The value of electronic laboratory notebooks in the management of data integrity and data availability |url=https://www.medra.org/servlet/aliasResolver?alias=iospress&doi=10.3233/ISU-170843 |journal=Information Services & Use |volume=37 |issue=3 |pages=355–359 |doi=10.3233/ISU-170843}}</ref> Most devices are equipped with a USB, RS232, or RS485 port, thus providing network connectivity. But the absence of a physical network necessitates the usage of, for example, flash drives for data transport. This increases the danger of manipulation or loss of data.<ref name=":5" /> Writing down data manually and then digitizing it later on a computer used for data storage and analysis is also not a good solution. The analog handwritten data is prone to error, not good to search through, only accessible from the analog laboratory notebook, and has limited reusability, in contrast to the FAIR data principles.<ref name=":5" /><ref name=":12">{{Cite journal |last=Gauglitz |first=Günter |date=2018-08 |title=Lab 4.0: SiLA or OPC UA |url=http://link.springer.com/10.1007/s00216-018-1192-6 |journal=Analytical and Bioanalytical Chemistry |language=en |volume=410 |issue=21 |pages=5093–5094 |doi=10.1007/s00216-018-1192-6 |issn=1618-2642}}</ref> After it has been digitized without human errors, it has all the advantages of digital data. In order to make data gathering faster, simpler, and more accurate, a complete transition to automated data capture has to be made.<ref name=":12" /> This includes data flow from a whole bioprocess, including sampling, sample preparation, measurement, and [[data analysis]].<ref name=":4" /> When transitioning from analog to digital data, it is important to take the laboratory users needs into consideration. While aiming to automatically store data following FAIR data principles, the end user who is analyzing it still needs to be able to access and work with it. | ||
===Standard communication protocols=== | ===Standard communication protocols=== | ||
While a lot of laboratory device vendors still offer their own proprietary software for device control | While a lot of laboratory device vendors still offer their own proprietary software for device control<ref name=":8" /><ref>{{Cite journal |last=Hinkel |first=Georg |last2=Kunert |first2=Jörg |last3=Meredith |first3=Jason |date=2023-10 |title=The Tecan SiLA2 SDK: A royalty-free, open-source framework to develop SiLA2 servers and clients |url=https://linkinghub.elsevier.com/retrieve/pii/S2472630323000444 |journal=SLAS Technology |language=en |volume=28 |issue=5 |pages=334–344 |doi=10.1016/j.slast.2023.07.001}}</ref>, researchers are agreeing that the future of digitalization in the laboratory lies in the common use of standardized device communication protocols.<ref name=":3" /><ref name=":5" /><ref name=":8" /><ref name=":12" /><ref name=":13">{{Citation |last=Seidel |first=Simon |last2=Cruz-Bournazou |first2=Mariano Nicolas |last3=Groß |first3=Sebastian |last4=Schollmeyer |first4=Julia Katharina |last5=Kurreck |first5=Anke |last6=Krauss |first6=Stefan |last7=Neubauer |first7=Peter |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=A Comprehensive IT Infrastructure for an Enzymatic Product Development in a Digitalized Biotechnological Laboratory |url=https://link.springer.com/10.1007/10_2022_207 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=61–82 |doi=10.1007/10_2022_207 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref><ref>{{Cite journal |last=Bai |first=Jiaru |last2=Cao |first2=Liwei |last3=Mosbach |first3=Sebastian |last4=Akroyd |first4=Jethro |last5=Lapkin |first5=Alexei A. |last6=Kraft |first6=Markus |date=2022-02-28 |title=From Platform to Knowledge Graph: Evolution of Laboratory Automation |url=https://pubs.acs.org/doi/10.1021/jacsau.1c00438 |journal=JACS Au |language=en |volume=2 |issue=2 |pages=292–309 |doi=10.1021/jacsau.1c00438 |issn=2691-3704 |pmc=PMC8889618 |pmid=35252980}}</ref><ref name=":14">{{Citation |last=Freundel |first=Matthias |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=Comparison of Laboratory Standards |url=https://link.springer.com/10.1007/10_2022_205 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=133–145 |doi=10.1007/10_2022_205 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref><ref name=":15">{{Cite journal |last=Wolf |first=Ádám |last2=Wolton |first2=David |last3=Trapl |first3=Josef |last4=Janda |first4=Julien |last5=Romeder-Finger |first5=Stefan |last6=Gatternig |first6=Thomas |last7=Farcet |first7=Jean-Baptiste |last8=Galambos |first8=Péter |last9=Széll |first9=Károly |date=2022-02 |title=Towards robotic laboratory automation Plug & Play: The “LAPP” framework |url=https://linkinghub.elsevier.com/retrieve/pii/S2472630321000224 |journal=SLAS Technology |language=en |volume=27 |issue=1 |pages=18–25 |doi=10.1016/j.slast.2021.11.003}}</ref><ref>{{Cite journal |last=Schmieder |first=Florian |last2=Polk |first2=Christoph |last3=Gottlöber |first3=Felix |last4=Schöps |first4=Patrick |last5=Sonntag |first5=Frank |last6=Deuse |first6=Ronny |last7=Jede |first7=Aline |last8=Petzold |first8=Thomas |date=2019-09-01 |title=Universal LIMS based platform for the automated processing of cell-based assays |url=https://www.degruyter.com/document/doi/10.1515/cdbme-2019-0110/html |journal=Current Directions in Biomedical Engineering |language=en |volume=5 |issue=1 |pages=437–439 |doi=10.1515/cdbme-2019-0110 |issn=2364-5504}}</ref><ref name=":16">{{Cite journal |last=Porr |first=Marc |last2=Marquard |first2=Daniel |last3=Stanislawski |first3=Nils |last4=Austerjost |first4=Jonas |last5=Russo |first5=Mario |last6=Bungers |first6=Simon |last7=Klimmt |first7=Christoph |last8=Scheper |first8=Thomas |last9=Beutel |first9=Sascha |last10=Lindner |first10=Patrick |date=2019-03 |title=smartLAB – Interaktives Arbeiten in digitalisierter Laborumgebung |url=https://onlinelibrary.wiley.com/doi/10.1002/cite.201800090 |journal=Chemie Ingenieur Technik |language=en |volume=91 |issue=3 |pages=285–293 |doi=10.1002/cite.201800090 |issn=0009-286X}}</ref><ref name=":17">{{Cite journal |last=Austerjost |first=Jonas |last2=Porr |first2=Marc |last3=Riedel |first3=Noah |last4=Geier |first4=Dominik |last5=Becker |first5=Thomas |last6=Scheper |first6=Thomas |last7=Marquard |first7=Daniel |last8=Lindner |first8=Patrick |last9=Beutel |first9=Sascha |date=2018-10 |title=Introducing a Virtual Assistant to the Lab: A Voice User Interface for the Intuitive Control of Laboratory Instruments |url=https://linkinghub.elsevier.com/retrieve/pii/S2472630322012651 |journal=SLAS Technology |language=en |volume=23 |issue=5 |pages=476–482 |doi=10.1177/2472630318788040}}</ref> To connect all laboratory devices to one digital infrastructure, it is aspirational for all devices to use the same standard communication protocol to achieve a plug-and-play environment in bioprocess laboratories.<ref name=":12" /> Device integration is faster, easier, and better to maintain when vendor-independent communication protocols are more common.<ref name=":18">{{Cite journal |last=Porr |first=Marc |last2=Lange |first2=Ferdinand |last3=Marquard |first3=Daniel |last4=Niemeyer |first4=Laura |last5=Lindner |first5=Patrick |last6=Scheper |first6=Thomas |last7=Beutel |first7=Sascha |date=2021-03 |title=Implementing a digital infrastructure for the lab using a central laboratory server and the SiLA2 communication standard |url=https://onlinelibrary.wiley.com/doi/10.1002/elsc.202000053 |journal=Engineering in Life Sciences |language=en |volume=21 |issue=3-4 |pages=208–219 |doi=10.1002/elsc.202000053 |issn=1618-0240 |pmc=PMC7923558 |pmid=33716619}}</ref> | ||
While offering device control with standard communication protocols might be costly for vendors in the beginning due to initial software development, it offers advantages as well. Using a communication standard as an instrument vendor will reduce the complexity of their software documentation since the basics have already been well documented. Thus, a detailed documentation of manufacturer-specific proprietary device drivers will become redundant. This simplification of software development can then reduce costs for device vendors. When using standard protocols for device communication, instrument vendors are contributing to the management of data according to the FAIR data principles, leaving them to focus on new innovations and features in their devices. Customers can then choose devices based upon those innovations instead of technical practicalities. Additionally, more companies and laboratories have the ability to integrate devices with standard communication protocols into their digital infrastructures because they are not limited by interface or platform incompatibilities. This will then lead to an increased purchase of consumables for those devices. | While offering device control with standard communication protocols might be costly for vendors in the beginning due to initial software development, it offers advantages as well. Using a communication standard as an instrument vendor will reduce the complexity of their software documentation since the basics have already been well documented. Thus, a detailed documentation of manufacturer-specific proprietary device drivers will become redundant. This simplification of software development can then reduce costs for device vendors. When using standard protocols for device communication, instrument vendors are contributing to the management of data according to the FAIR data principles, leaving them to focus on new innovations and features in their devices. Customers can then choose devices based upon those innovations instead of technical practicalities. Additionally, more companies and laboratories have the ability to integrate devices with standard communication protocols into their digital infrastructures because they are not limited by interface or platform incompatibilities. This will then lead to an increased purchase of consumables for those devices.<ref name=":19">{{Citation |last=Juchli |first=Daniel |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=SiLA 2: The Next Generation Lab Automation Standard |url=https://link.springer.com/10.1007/10_2022_204 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=147–174 |doi=10.1007/10_2022_204 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref><ref>{{Cite journal |last=Ktori, S. |date=May 2021 |title=SiLA for Instrument Vendors |url=https://sila-standard.com/wp-content/uploads/2021/06/SiLA-for-instrument-vendors-WhitePaper.pdf |journal=SiLA Analyzed |volume=1 |pages=1–4}}</ref><ref name=":20">{{Cite web |title=SiLA Frequently Asked Questions |url=https://sila-standard.com/faq/ |publisher=SiLA Consortium |accessdate=02 September 2023}}</ref><ref>{{Cite web |title=Unified Architecture |url=https://opcfoundation.org/about/opc-technologies/opc-ua/ |publisher=OPC Foundation}}</ref><ref name=":21">{{Cite journal |last=Bär |first=Henning |last2=Hochstrasser |first2=Remo |last3=Papenfuß |first3=Bernd |date=2012-04 |title=SiLA: Basic Standards for Rapid Integration in Laboratory Automation |url=https://linkinghub.elsevier.com/retrieve/pii/S2472630322016673 |journal=SLAS Technology |language=en |volume=17 |issue=2 |pages=86–95 |doi=10.1177/2211068211424550}}</ref> Agreeing on physical standards like the [[microtiter plate]] was an advantage for everyone in the end.<ref name=":14" /> Using standardized tools and parts in the production process is decreasing costs and complexity due to purchasing larger quantities of the same part rather than needing multiple specific parts. The earlier standards are getting adopted and implemented in the working process the more cost efficient it will be. Manufacturers are also challenged to differentiate their product from other manufacturers in other areas than compatibility. This leads to a better competition and an overall increase in developing new technologies rather than sticking to proprietary solutions.<ref name=":14" /><ref>{{Cite journal |last=Rachuri |first=Sudarsan |last2=Subrahmanian |first2=Eswaran |last3=Bouras |first3=Abdelaziz |last4=Fenves |first4=Steven J. |last5=Foufou |first5=Sebti |last6=Sriram |first6=Ram D. |date=2008-07 |title=Information sharing and exchange in the context of product lifecycle management: Role of standards |url=https://linkinghub.elsevier.com/retrieve/pii/S0010448507001613 |journal=Computer-Aided Design |language=en |volume=40 |issue=7 |pages=789–800 |doi=10.1016/j.cad.2007.06.012}}</ref> For consumers on the other hand, standardization will help to easily maintain devices even if the manufacturer no longer exists. Standards need to be flexible enough to go beyond their intended purpose or otherwise it will limit their use. | ||
Before implementing a communication standard, a holistic plan needs to be made from scientists in cooperation with IT experts. Laboratory work needs to be simplified, and the scientist needs to be supported through standard device connectivity. Users need to have a clear benefit from using a standard, otherwise digitalization efforts will not be rewarded by regular use. As long as the use of proprietary device control software is more convenient, the use of standard communication protocols is ineffective. These benefits can, for example, be achieved by automating parts of the workflow that would otherwise result in tedious manual work, like data acquisition and central data storage following FAIR data principles. Other means to support the user in the laboratory are semi-automated SOPs, where device parameters are already preset after digitally selecting the required workflow. Using a standard protocol for device communication can realize these aspects when all devices are connected by a central device control server, as demonstrated by Porr ''et al.'' | Before implementing a communication standard, a holistic plan needs to be made from scientists in cooperation with IT experts. Laboratory work needs to be simplified, and the scientist needs to be supported through standard device connectivity. Users need to have a clear benefit from using a standard, otherwise digitalization efforts will not be rewarded by regular use. As long as the use of proprietary device control software is more convenient, the use of standard communication protocols is ineffective. These benefits can, for example, be achieved by automating parts of the workflow that would otherwise result in tedious manual work, like data acquisition and central data storage following FAIR data principles. Other means to support the user in the laboratory are semi-automated SOPs, where device parameters are already preset after digitally selecting the required workflow. Using a standard protocol for device communication can realize these aspects when all devices are connected by a central device control server, as demonstrated by Porr ''et al.''<ref name=":18" /> and Rachuri ''et al.''<ref name=":22">{{Cite journal |last=Bromig |first=Lukas |last2=Leiter |first2=David |last3=Mardale |first3=Alexandru-Virgil |last4=von den Eichen |first4=Nikolas |last5=Bieringer |first5=Emmeran |last6=Weuster-Botz |first6=Dirk |date=2022-01 |title=The SiLA 2 Manager for rapid device integration and workflow automation |url=https://linkinghub.elsevier.com/retrieve/pii/S2352711022000103 |journal=SoftwareX |language=en |volume=17 |pages=100991 |doi=10.1016/j.softx.2022.100991}}</ref> After successfully choosing and implementing standard communication, it is important to keep the infrastructure open to new device additions and upgrades without being a burden on laboratory workers. Two standard communication protocols are currently coexisting in bioprocess development laboratories: Standardization in Lab Automation 2 (SiLA2) and the Laboratory and Analytical Device Standard (LADS), based on Open Platform Communications Unified Architecture (OPC UA).<ref name=":8" /><ref name=":19" /><ref name=":20" /><ref name=":21" /><ref name=":23">{{Citation |last=Brendel |first=Alexander |last2=Dorfmüller |first2=Franziska |last3=Liebscher |first3=Albrecht |last4=Kraus |first4=Patrick |last5=Kress |first5=Kai |last6=Oehme |first6=Heiko |last7=Arnold |first7=Matthias |last8=Koschitzki |first8=Robert |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=Laboratory and Analytical Device Standard (LADS): A Communication Standard Based on OPC UA for Networked Laboratories |url=https://link.springer.com/10.1007/10_2022_209 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=175–194 |doi=10.1007/10_2022_209 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref><ref name=":24">{{Cite web |date=2023 |title=Was ist OPC UA? Die wichtigsten Begriffe im Überblick |work=OPC Router |url=https://www.opc-router.de/was-ist-opc-ua/ |publisher=inray Industriesoftware GmbH}}</ref> | ||
====SiLA2==== | ====SiLA2==== | ||
“SiLA's mission is to establish international standards which create open connectivity in lab automation, to enable lab digitalization.” | “SiLA's mission is to establish international standards which create open connectivity in lab automation, to enable lab digitalization.”<ref name=":20" /> This open-source communication standard is based on a client-server infrastructure where every laboratory device is a SiLA2 server that offers a set of features. SiLA2 servers advertise themselves on the network and are discovered by SiLA2 clients like a [[laboratory information management system]] (LIMS) or the SiLA2 Manager developed by Bromig ''et al.''<ref name=":19" /><ref name=":21" /><ref name=":22" /> The clients recognize the offered features by the servers and can use or call them. Real-time observation of parameters is possible through the subscription to observable properties. For integration of legacy devices, Porr ''et al.'' developed a gateway module to enable SiLA communication.<ref name=":25">{{Cite journal |last=Porr |first=Marc |last2=Schwarz |first2=Sebastian |last3=Lange |first3=Ferdinand |last4=Niemeyer |first4=Laura |last5=Hentrop |first5=Thorleif |last6=Marquard |first6=Daniel |last7=Lindner |first7=Patrick |last8=Scheper |first8=Thomas |last9=Beutel |first9=Sascha |date=2020-10 |title=Bringing IoT to the Lab: SiLA2 and Open-Source-Powered Gateway Module for Integrating Legacy Devices into the Digital Laboratory |url=https://linkinghub.elsevier.com/retrieve/pii/S2468067220300274 |journal=HardwareX |language=en |volume=8 |pages=e00118 |doi=10.1016/j.ohx.2020.e00118 |pmc=PMC9041174 |pmid=35498267}}</ref> On the software side, the gateway module has the advantage of clearly separating the tasks while still using a standard protocol. This plays a major role in scalability and maintainability. SiLA2 has been implemented in different frameworks using Java, C#, Python, or C++ for fast and easy integration of laboratory devices.<ref name=":22" /><ref name=":25" /><ref>{{Cite web |title=SiLA Downloads |url=https://sila-standard.com/downloads/ |publisher=SiLA Consortium |accessdate=14 April 2023}}</ref> | ||
====LADS based on OPC UA==== | ====LADS based on OPC UA==== | ||
OPC UA is a standard in multiple industries for data exchange between devices independent from the device vendor. | OPC UA is a standard in multiple industries for data exchange between devices independent from the device vendor.<ref name=":24" /> Just like SiLA2, OPC UA has a client-server infrastructure. OPC clients can be connected to any OPC server that is able to collect and further process provided data from the client. The previous version, OPC Classic, was limited to the operating system Microsoft Windows; however, OPC UA is platform-independent. OPC was used as an established standard in industry even before the need for a standard communication protocol for laboratory equipment manifested itself.<ref name=":24" /> The problem of transferring this digital communication standard from industry to bioprocess development research laboratories is the added flexibility and heterogeneity of devices that make those laboratories a lot more complex than industrial production processes when looking at the digitalization part.<ref name=":23" /> This is why a working group from the industrial association Spectaris developed the communication protocol LADS based on OPC UA.<ref name=":8" /><ref>{{Cite web |date=December 2020 |title=SPECTARIS Standard for Laboratory Equipment Communication |work=OPC Connect |url=https://opcconnect.opcfoundation.org/2020/12/spectaris-standard-for-laboratory-equipment-communication/ |publisher=OPC Foundation}}</ref> “The goal of LADS is to create a manufacturer-independent, open standard for analytical and laboratory equipment.”<ref>{{Cite web |title=LADS – Laboratory and Analytical Device Standard |url=https://opcfoundation.org/markets-collaboration/lads/ |publisher=OPC Foundation}}</ref> OPC UA provides a way to integrate legacy devices into the digital infrastructure by providing wrappers for existing software.<ref name=":26">{{Cite book |last=Mahnke |first=Wolfgang |last2=Leitner |first2=Stefan-Helmut |last3=Damm |first3=Matthias |date=2009 |title=OPC Unified Architecture |url=http://link.springer.com/10.1007/978-3-540-68899-0 |language=en |publisher=Springer Berlin Heidelberg |place=Berlin, Heidelberg |doi=10.1007/978-3-540-68899-0 |isbn=978-3-540-68898-3}}</ref> Implementing OPC UA is possible in different programming languages such as Java, Python, C++, or C#.<ref name=":26" /> OPC UA is not just enabling digital connectivity inside the laboratory but also connecting research laboratories to industrial infrastructures due to OPC UA already being established in industrial environments.<ref name=":8" /> | ||
===Digital twins=== | ===Digital twins=== | ||
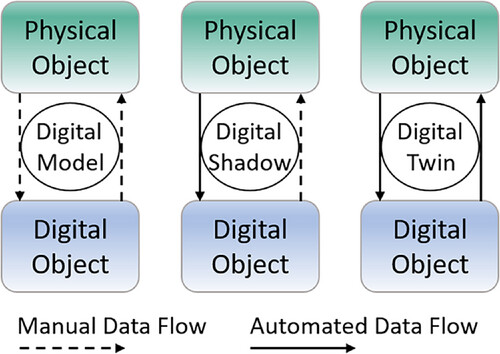
Through the development of Industry 4.0 and progressing [[internet of things]] (IoT) connectivity, the number of digital twins is increasing also in the environment of bioprocess engineering laboratories. IoT in this context describes the idea to connect physical objects like laboratory devices to a network or the internet to ensure status and information processing, as well as the ability to communicate and send and receive data. | Through the development of Industry 4.0 and progressing [[internet of things]] (IoT) connectivity, the number of digital twins is increasing also in the environment of bioprocess engineering laboratories. IoT in this context describes the idea to connect physical objects like laboratory devices to a network or the internet to ensure status and information processing, as well as the ability to communicate and send and receive data.<ref name=":13" /><ref name=":27">{{Cite journal |last=Poongothai |first=M. |last2=Subramanian |first2=P. Muthu |last3=Rajeswari |first3=A. |date=2018-04 |title=Design and implementation of IoT based smart laboratory |url=https://ieeexplore.ieee.org/document/8387090/ |journal=2018 5th International Conference on Industrial Engineering and Applications (ICIEA) |publisher=IEEE |place=Singapore |pages=169–173 |doi=10.1109/IEA.2018.8387090 |isbn=978-1-5386-5747-8}}</ref><ref>{{Cite journal |last=Patel, K.K; Patel, S.M. |year=2016 |title=Internet of Things-IOT: Definition, Characteristics, Architecture, Enabling Technologies, Application & Future Challenges |url=https://fardapaper.ir/mohavaha/uploads/2019/09/Fardapaper-Internet-of-Things-IOT-Definition-Characteristics-Architecture-Enabling-Technologies-Application-Future-Challenges.pdf |journal=International Journal of Engineering Science and Computing |volume=6 |issue=5 |pages=6122–31 |doi=10.4010/2016.1482}}</ref><ref name=":28">{{Cite journal |last=Garg |first=Hittu |last2=Dave |first2=Mayank |date=2019-04 |title=Securing IoT Devices and SecurelyConnecting the Dots Using REST API and Middleware |url=https://ieeexplore.ieee.org/document/8777334/ |journal=2019 4th International Conference on Internet of Things: Smart Innovation and Usages (IoT-SIU) |publisher=IEEE |place=Ghaziabad, India |pages=1–6 |doi=10.1109/IoT-SIU.2019.8777334 |isbn=978-1-7281-1253-4}}</ref><ref name=":29">{{Citation |last=Söldner |first=Robert |last2=Rheinländer |first2=Sophia |last3=Meyer |first3=Tim |last4=Olszowy |first4=Michael |last5=Austerjost |first5=Jonas |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=Human–Device Interaction in the Life Science Laboratory |url=https://link.springer.com/10.1007/10_2021_183 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=83–113 |doi=10.1007/10_2021_183 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref> This omnipresent keyword "[[digital twin]]" is described by Fuller ''et al.'' as the “effortless integration of data between a physical and virtual machine in either direction.”<ref name=":30">{{Cite journal |last=Fuller |first=Aidan |last2=Fan |first2=Zhong |last3=Day |first3=Charles |last4=Barlow |first4=Chris |date=2020 |title=Digital Twin: Enabling Technologies, Challenges and Open Research |url=https://ieeexplore.ieee.org/document/9103025/ |journal=IEEE Access |volume=8 |pages=108952–108971 |doi=10.1109/ACCESS.2020.2998358 |issn=2169-3536}}</ref> This concept is an invaluable advancement in bioprocess development, whereas a differentiation between "digital models," "digital shadows," and "digital twins" is important. The difference between these, as depicted in Figure 1, are the manual and automated data flows between the physical and digital object. All three concepts have a physical and digital object that are connected by data flow. Within the digital model, the data in transported manually between the physical and digital object. The digital shadow has automated data flow from the physical to the digital object, but manual data flow from the digital to the physical object. A digital twin, however, has automated data flow in both directions. Thus, only when using a digital twin, changes in the physical object will result in changes in the digital object and vice versa.<ref name=":3" /><ref name=":30" /><ref name=":31">{{Cite journal |last=Kritzinger |first=Werner |last2=Karner |first2=Matthias |last3=Traar |first3=Georg |last4=Henjes |first4=Jan |last5=Sihn |first5=Wilfried |date=2018 |title=Digital Twin in manufacturing: A categorical literature review and classification |url=https://linkinghub.elsevier.com/retrieve/pii/S2405896318316021 |journal=IFAC-PapersOnLine |language=en |volume=51 |issue=11 |pages=1016–1022 |doi=10.1016/j.ifacol.2018.08.474}}</ref> | ||
| Line 68: | Line 62: | ||
{| border="0" cellpadding="5" cellspacing="0" width="500px" | {| border="0" cellpadding="5" cellspacing="0" width="500px" | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 1.''' Portrayal of differences in the data flow of digital models, shadows and twins. Figure based on Richter ''et al.'' | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 1.''' Portrayal of differences in the data flow of digital models, shadows and twins. Figure based on Richter ''et al.''<ref name=":3" />, Fuller ''et al.''<ref name=":30" />, and Kritzinger ''et al.''<ref name=":31" /></blockquote> | ||
|- | |- | ||
|} | |} | ||
|} | |} | ||
Major challenges in the process of implementing digital twins in the bioprocess laboratory are [[Information security|data security]] and [[Information privacy|privacy]] concerns, device connectivity, trust issues regarding the use of technology, and development of a laboratory-wide digital infrastructure, including data storage space. Difficult to predict and complex processes especially in the biological laboratory complicate the development of digital twins as well. The predominant benefits on the other hand include automated data (and metadata) generation, processing, and analyzing of those as well as data traceability. All this is contributing to the aforementioned FAIR data principles. Laboratories that use digital tracking of consumables in their digital twins are able to automate their ordering system making manual inventory obsolete. Therefore, digital twins are the key technology to a new and transformed laboratory work, with the potential to automated knowledge generation. | Major challenges in the process of implementing digital twins in the bioprocess laboratory are [[Information security|data security]] and [[Information privacy|privacy]] concerns, device connectivity, trust issues regarding the use of technology, and development of a laboratory-wide digital infrastructure, including data storage space. Difficult to predict and complex processes especially in the biological laboratory complicate the development of digital twins as well. The predominant benefits on the other hand include automated data (and metadata) generation, processing, and analyzing of those as well as data traceability. All this is contributing to the aforementioned FAIR data principles. Laboratories that use digital tracking of consumables in their digital twins are able to automate their ordering system making manual inventory obsolete. Therefore, digital twins are the key technology to a new and transformed laboratory work, with the potential to automated knowledge generation.<ref name=":3" /><ref name=":31" /> | ||
===Interaction with laboratory devices=== | ===Interaction with laboratory devices=== | ||
Besides using standardized device communication protocols, full digitalization cannot be achieved without considering the end user. Digital solutions assisting the laboratory worker but also interacting with devices should therefore be straightforward and convenient to use. The device interactions with laboratory equipment in bioprocess development laboratories has changed over the last years, making it more efficient and intuitive with every additional solution, providing more interaction possibilities. | Besides using standardized device communication protocols, full digitalization cannot be achieved without considering the end user. Digital solutions assisting the laboratory worker but also interacting with devices should therefore be straightforward and convenient to use. The device interactions with laboratory equipment in bioprocess development laboratories has changed over the last years, making it more efficient and intuitive with every additional solution, providing more interaction possibilities.<ref name=":32">{{Cite journal |last=Wolf |first=Adam |last2=Galambos |first2=Peter |last3=Szell |first3=Karoly |date=2020-07 |title=Device Integration Concepts in Laboratory Automation |url=https://ieeexplore.ieee.org/document/9147171/ |journal=2020 IEEE 24th International Conference on Intelligent Engineering Systems (INES) |publisher=IEEE |place=Reykjavík, Iceland |pages=171–178 |doi=10.1109/INES49302.2020.9147171 |isbn=978-1-7281-1059-2}}</ref> | ||
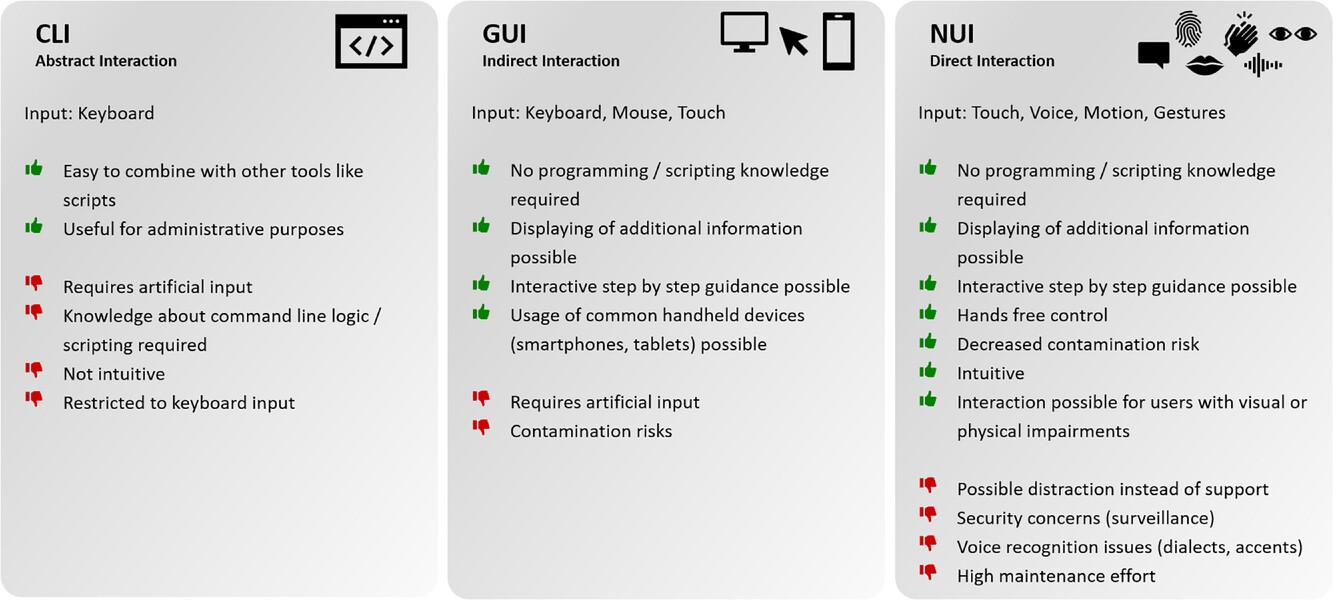
The use of laboratory devices itself is supposed to not interfere with the experiment, slow it down, or complicate it. Consequently, device operation should be as intuitive as possible. There are three different interactions levels for device control. Starting at the lowest level, there is abstract interaction, using command line interfaces (CLI), followed by the next level, the indirect interaction, using graphical user interfaces (GUI). The last and most intuitive level of device interaction is the direct interaction using natural user interfaces (NUI). | The use of laboratory devices itself is supposed to not interfere with the experiment, slow it down, or complicate it. Consequently, device operation should be as intuitive as possible. There are three different interactions levels for device control. Starting at the lowest level, there is abstract interaction, using command line interfaces (CLI), followed by the next level, the indirect interaction, using graphical user interfaces (GUI). The last and most intuitive level of device interaction is the direct interaction using natural user interfaces (NUI). | ||
CLIs have the advantage of keyboard input from the user and are easy to integrate or combine with other tools like scripts. However, using CLIs as the interaction medium within the laboratory environment requires basic knowledge of command line logic and scripting. This skill cannot be expected from everyone working in the laboratory due to their different educational backgrounds. One approach to accelerate the transformation towards digitalized laboratories is the addition of basic IT classes (e.g., for basic scripting knowledge) to biotechnological education. However, CLIs as the only device interface are not a suitable solution for general and intuitive device interaction. Yet additional CLI device interaction may prove useful for administrative purposes or for integrating supplementary software. | CLIs have the advantage of keyboard input from the user and are easy to integrate or combine with other tools like scripts. However, using CLIs as the interaction medium within the laboratory environment requires basic knowledge of command line logic and scripting. This skill cannot be expected from everyone working in the laboratory due to their different educational backgrounds. One approach to accelerate the transformation towards digitalized laboratories is the addition of basic IT classes (e.g., for basic scripting knowledge) to biotechnological education. However, CLIs as the only device interface are not a suitable solution for general and intuitive device interaction. Yet additional CLI device interaction may prove useful for administrative purposes or for integrating supplementary software.<ref name=":32" /> | ||
The next level of interaction with laboratory devices is the indirect interaction using GUIs. This has the advantage that no programming or scripting knowledge is needed for device interaction. Thus, all people in the laboratory are able to operate devices equipped with a GUI. Additionally, more information can be displayed for the user on the screen while mouse and keyboard input is enabled. A lot of devices have a vendor-specific and often proprietary control software. This often runs on a desktop PC positioned next to the device inside the laboratory. Modern devices, however, do not only have the possibility of local interaction but also offer remote interaction via network connection. Those GUIs are then deployed as touch-based network-like web interfaces, also allowing the usage of handheld devices as the end device. These handheld devices like smartphones, tablets, or touch beamers have the advantage that people are used to operating touch displays from their personal life making device interaction even more intuitive. Additionally, the use of mobile applications on smartphones and tablets is enabled. | The next level of interaction with laboratory devices is the indirect interaction using GUIs. This has the advantage that no programming or scripting knowledge is needed for device interaction. Thus, all people in the laboratory are able to operate devices equipped with a GUI. Additionally, more information can be displayed for the user on the screen while mouse and keyboard input is enabled. A lot of devices have a vendor-specific and often proprietary control software. This often runs on a desktop PC positioned next to the device inside the laboratory. Modern devices, however, do not only have the possibility of local interaction but also offer remote interaction via network connection. Those GUIs are then deployed as touch-based network-like web interfaces, also allowing the usage of handheld devices as the end device. These handheld devices like smartphones, tablets, or touch beamers have the advantage that people are used to operating touch displays from their personal life making device interaction even more intuitive. Additionally, the use of mobile applications on smartphones and tablets is enabled.<ref name=":33">{{Cite journal |last=Austerjost |first=Jonas |last2=Marquard |first2=Daniel |last3=Raddatz |first3=Lukas |last4=Geier |first4=Dominik |last5=Becker |first5=Thomas |last6=Scheper |first6=Thomas |last7=Lindner |first7=Patrick |last8=Beutel |first8=Sascha |date=2017-08 |title=A smart device application for the automated determination of E. coli colonies on agar plates |url=https://onlinelibrary.wiley.com/doi/10.1002/elsc.201700056 |journal=Engineering in Life Sciences |language=en |volume=17 |issue=8 |pages=959–966 |doi=10.1002/elsc.201700056 |issn=1618-0240 |pmc=PMC6999497 |pmid=32624845}}</ref><ref>{{Cite journal |last=Gan |first=Samuel Ken-En |last2=Poon |first2=Jun-Kai |date=2016-12 |title=The world of biomedical apps: their uses, limitations, and potential |url=http://scientificphoneapps.springeropen.com/articles/10.1186/s41070-016-0009-2 |journal=Scientific Phone Apps and Mobile Devices |language=en |volume=2 |issue=1 |pages=6 |doi=10.1186/s41070-016-0009-2 |issn=2364-4958}}</ref><ref>{{Cite journal |last=Min |first=Hyun Jung |last2=Mina |first2=Hansel A. |last3=Deering |first3=Amanda J. |last4=Bae |first4=Euiwon |date=2021-09 |title=Development of a smartphone-based lateral-flow imaging system using machine-learning classifiers for detection of Salmonella spp. |url=https://linkinghub.elsevier.com/retrieve/pii/S0167701221001561 |journal=Journal of Microbiological Methods |language=en |volume=188 |pages=106288 |doi=10.1016/j.mimet.2021.106288}}</ref> | ||
The use of touch-based interaction media is introducing the next level of device interaction: NUI. Besides touch-based interaction media, NUIs also include voice user interfaces (VUI) and interaction via gestures and motions. Artificial input like keyboard and mouse input becomes obsolete. This direct interaction with laboratory devices is leading to a simplified and even more intuitive way of working in the digitalized laboratory environment. Even though NUIs have a lot of advantages (Figure 2) they are not undisputed due to security concerns or voice recognition issues. | The use of touch-based interaction media is introducing the next level of device interaction: NUI. Besides touch-based interaction media, NUIs also include voice user interfaces (VUI) and interaction via gestures and motions. Artificial input like keyboard and mouse input becomes obsolete. This direct interaction with laboratory devices is leading to a simplified and even more intuitive way of working in the digitalized laboratory environment. Even though NUIs have a lot of advantages (Figure 2) they are not undisputed due to security concerns or voice recognition issues.<ref>{{Cite journal |last=Tatman |first=Rachael |last2=Kasten |first2=Conner |date=2017-08-20 |title=Effects of Talker Dialect, Gender & Race on Accuracy of Bing Speech and YouTube Automatic Captions |url=https://www.isca-archive.org/interspeech_2017/tatman17_interspeech.html |journal=Interspeech 2017 |language=en |publisher=ISCA |pages=934–938 |doi=10.21437/Interspeech.2017-1746}}</ref><ref>{{Cite journal |last=Salazar Ch. |first=Gustavo D. |last2=Venegas |first2=Carlos |last3=Baca |first3=Michael |last4=Rodriguez |first4=Ismael |last5=Marrone |first5=Luis |date=2018-11 |title=Open Middleware proposal for IoT focused on Industry 4.0 |url=https://ieeexplore.ieee.org/document/8588117/ |journal=2018 IEEE 2nd Colombian Conference on Robotics and Automation (CCRA) |publisher=IEEE |place=Barranquilla |pages=1–6 |doi=10.1109/CCRA.2018.8588117 |isbn=978-1-5386-8464-1}}</ref> Austerjost ''et al.'' showed in their work that VUIs can enable hands-free device control, which can be particularly advantageous in [[microbiology]] laboratories regarding contamination risks.<ref name=":17" /> The topic of human-device interaction is described in further detail in the work of Söldner ''et al.''<ref name=":29" /> | ||
| Line 99: | Line 93: | ||
==Digitalization strategies== | ==Digitalization strategies== | ||
The way digitalization in laboratories looks can be very different everywhere. Bioprocess development is still relying on experienced workers doing experiments in the laboratory and analyzing data afterwards. Digitalization with integrating devices, digital and physical assistants, automation, and simulation is setting a new direction for laboratory work. | The way digitalization in laboratories looks can be very different everywhere. Bioprocess development is still relying on experienced workers doing experiments in the laboratory and analyzing data afterwards. Digitalization with integrating devices, digital and physical assistants, automation, and simulation is setting a new direction for laboratory work.<ref name=":1" /><ref name=":34">{{Cite journal |last=Biermann |first=Ferdinand |last2=Mathews |first2=Julius |last3=Nießing |first3=Bastian |last4=König |first4=Niels |last5=Schmitt |first5=Robert H. |date=2021-05-29 |title=Automating Laboratory Processes by Connecting Biotech and Robotic Devices—An Overview of the Current Challenges, Existing Solutions and Ongoing Developments |url=https://www.mdpi.com/2227-9717/9/6/966 |journal=Processes |language=en |volume=9 |issue=6 |pages=966 |doi=10.3390/pr9060966 |issn=2227-9717}}</ref> One digitalization strategy alone will not be able to meet all needs of the various laboratories that exists in the academic bioprocess development field.<ref name=":14" /> In the following subsections, different approaches of digitalization with their respective applications are presented and discussed. | ||
===Digital integration of laboratory devices=== | ===Digital integration of laboratory devices=== | ||
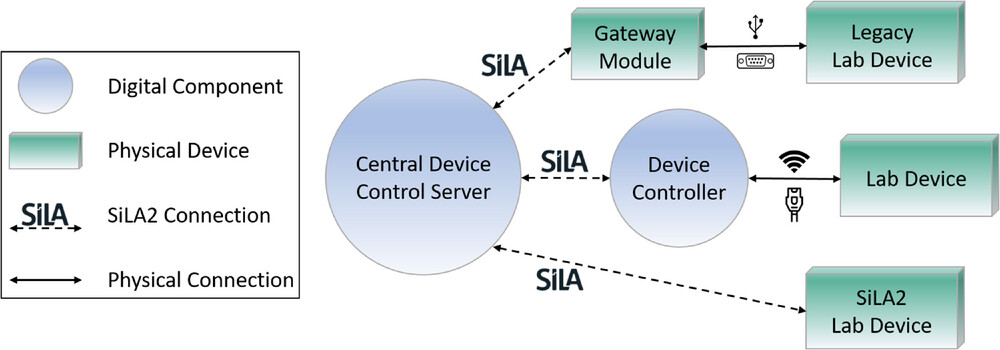
The first step to digitalize any laboratory is to connect devices to a digital infrastructure. This connection can look different for each laboratory, depending on their individual needs. Due to high topic variance in some academic research laboratories, the devices are usually waiting to be used by a researcher in their required manual way. | The first step to digitalize any laboratory is to connect devices to a digital infrastructure. This connection can look different for each laboratory, depending on their individual needs. Due to high topic variance in some academic research laboratories, the devices are usually waiting to be used by a researcher in their required manual way.<ref name=":34" /> Devices in these laboratories are described by Porr ''et al.'' as a “craftsman's toolbox.”<ref name=":25" /> Integrating them into a highly automated workflow might hinder research and flexibility, which is why there are different approaches to hardware integration in academic research laboratories. Laboratory devices can be divided into three categories with different levels of technical prerequisites. In order to connect or control any device with a digital infrastructure, a central device control server needs to be developed first. The SiLA2 manager developed by Bromig ''et al.''<ref name=":22" /> or the DeviceLayer from Porr ''et al.''<ref name=":18" /> are examples of this. The connection to different laboratory devices will then look different due to their category. While legacy laboratory devices do not have internet access connection points but rather a USB or other serial connection ports (like RS232), they first need to be connected to a physical device that bridges the connection to the network as well as to the central control server and enables standard communication. This can be achieved using a gateway module. Laboratory devices with network access (WLAN or Ethernet) do not need such a physical device in order to be connected using a standard device communication protocol. A simple digital component like a device controller is sufficient to integrate that device into the digital infrastructure. Laboratory devices that come with a standard driver like SiLA2 or OPC UA can be controlled directly from the central control server.<ref name=":18" /> Figure 3 shows this concept with SiLA2 as the example for the standard device communication protocol. | ||
| Line 116: | Line 110: | ||
|} | |} | ||
For the integration of legacy laboratory devices, Porr ''et al.'' developed a gateway module to enable SiLA-based communication. This gateway module is based on an embedded computer that runs a Linux image. The legacy device is connected to this module via USB or RS232 and a SiLA2 server on the other side that can control the device hosted on the gateway module. Thus, communication using the SiLA communication protocol is enabled. In their work, they showed how to use this connection device to integrate a magnetic stirrer into the digital infrastructure. | For the integration of legacy laboratory devices, Porr ''et al.'' developed a gateway module to enable SiLA-based communication. This gateway module is based on an embedded computer that runs a Linux image. The legacy device is connected to this module via USB or RS232 and a SiLA2 server on the other side that can control the device hosted on the gateway module. Thus, communication using the SiLA communication protocol is enabled. In their work, they showed how to use this connection device to integrate a magnetic stirrer into the digital infrastructure.<ref name=":25" /> Bromig ''et al.'' also connected their laboratory devices using the SiLA2 communication protocol in their work. They developed a SiLA2 Manager that is able to discover, connect to and control SiLA2 drivers generated with SiLA2 Python. Using this software framework, they connected various laboratory devices needed for their use case cultivation workflow with intermittent glucose feeding.<ref name=":22" /> | ||
Schmid ''et al.'' used the iLAB software framework (meanwhile known as zenLAB from infoteam | Schmid ''et al.'' used the iLAB software framework (meanwhile known as zenLAB from infoteam<ref>{{Cite web |title=Life Science |url=https://infoteam.de/unsere-maerkte/life-science |publisher=Inforteam Software}}</ref>) as a basis to develop their own software framework called iLAB-Bio. The iLAB software framework is able to communicate with SiLA and OPC interface protocols generating their own generic driver for each device. The communication with both emerging standard protocols on the market (SiLA and OPC) is making it a good [[middleware]] framework for a digitalized laboratory. Using this framework, they were able to integrate a liquid handling platform using SiLA as the device communication protocol. On top of that, they used SiLA-based communication to integrate a bioreactor system with forty-eight 10 mL reaction vessels for parallelization and miniaturization. They also integrated the design of experiment (DoE) software MODDE into their digital infrastructure. For this, they used a combination of SiLA-based communication and manually matching process data to MODDE responses. While this method includes proprietary data formats, it still improves the data exchange with the software, decreasing the possibilities of data loss or damage.<ref name=":5" /> | ||
Seidel ''et al.'' and Poongothai ''et al''. used various communication protocols (like SiLA2, OPC UA, gRPC, MQTT, or REST) to integrate laboratory devices into their comprehensive IT platform in an IoT fashion. Integrating devices using different interface options increases the maintenance cost and time, as well as the complexity, but this also allows the infrastructure to stay as flexible as possible. Automatically storing all available data in one place saves time, ensures data transparency, and reduces human error. | Seidel ''et al.'' and Poongothai ''et al''. used various communication protocols (like SiLA2, OPC UA, gRPC, MQTT, or REST) to integrate laboratory devices into their comprehensive IT platform in an IoT fashion. Integrating devices using different interface options increases the maintenance cost and time, as well as the complexity, but this also allows the infrastructure to stay as flexible as possible. Automatically storing all available data in one place saves time, ensures data transparency, and reduces human error.<ref name=":13" /> | ||
Austerjost ''et al.'' also used the concept of IoT while using proprietary commands for laboratory device control. | Austerjost ''et al.'' also used the concept of IoT while using proprietary commands for laboratory device control.<ref name=":17" /> They generated a digital device shadow for each device which was synchronized with the real laboratory device using the [[Cloud computing|cloud-based]] IoT broker AWS IoT. The modular device integration was realized using the programming tool Node-RED<ref>{{Cite web |title=Node-RED |url=https://nodered.org/ |publisher=OpenJS Foundation}}</ref>, deployed on a Windows PC with both devices serially connected to the PC. The serial communication was possible with proprietary port commands that were provided by the vendor. The modular setup allows more device integration in the future, but using proprietary elements in the digitalization process will not lead to a universal solution for digitalized laboratories.<ref name=":12" /><ref name=":17" /><ref name=":23" /> | ||
In 2021, Porr ''et al.'' developed a digital infrastructure where device integration was achieved using SiLA-based communication. | In 2021, Porr ''et al.'' developed a digital infrastructure where device integration was achieved using SiLA-based communication.<ref name=":18" /> The underlying concept of this solution was already published by Porr ''et al.'' in 2019.<ref name=":16" /> Ethernet, USB, and RS232 is used to connect the (legacy) devices<ref name=":25" /> with a wire making digital communication more stable than wireless communication due to less signal interfering possibilities. The infrastructure is based on a server-client-logic. It includes a process control system as their central process intelligence. Here, workflows, the sequence of steps required to complete the bioprocess, are planned and controlled. The process control system is connected to the aforementioned DeviceLayer, which communicates with the laboratory devices. After creating a workflow with the process control system using an intuitive GUI, the user can be guided through the laboratory work with the help of smartglasses as a NUI. The smartglasses show, for example, the next step of a complex assay that was incorporated into the workflow. Here it is important that this additional information is helping rather than distracting the user.<ref name=":16" /> The use of smartglasses is just one example of using assistive devices in the laboratory, which will be further examined in the next subsection. | ||
===Laboratory assistants=== | ===Laboratory assistants=== | ||
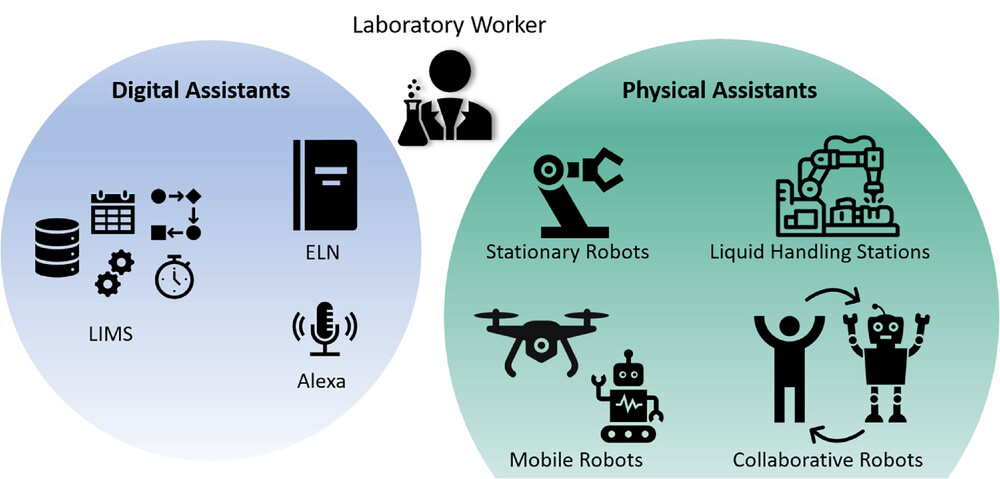
Next to digitalizing laboratory devices and thus enabling digital communication, there are also other means for digital assistance in the laboratory. They can be distinguished into digital and physical assistants (Figure 4), but for both groups, their main goal is to help and support the person working in the laboratory. When working with laboratory assistants, it is unavoidable to address issues like privacy and confidentiality beforehand. [[Encryption|Encrypted]] connections are an important factor for secure file transport. Other security methods like authentication, usage of (voice-)fingerprints, logging of system activities, message signing, or [[version control]] are becoming equally relevant when working with digital and physical assistants in digitalized laboratories. | Next to digitalizing laboratory devices and thus enabling digital communication, there are also other means for digital assistance in the laboratory. They can be distinguished into digital and physical assistants (Figure 4), but for both groups, their main goal is to help and support the person working in the laboratory. When working with laboratory assistants, it is unavoidable to address issues like privacy and confidentiality beforehand. [[Encryption|Encrypted]] connections are an important factor for secure file transport. Other security methods like authentication, usage of (voice-)fingerprints, logging of system activities, message signing, or [[version control]] are becoming equally relevant when working with digital and physical assistants in digitalized laboratories.<ref name=":13" /><ref name=":17" /><ref name=":19" /><ref name=":23" /><ref name=":27" /><ref name=":28" /><ref name=":33" /><ref name=":35">{{Cite journal |last=Prasad |first=Poonam J. |last2=Bodhe |first2=G.L. |date=2012-08 |title=Trends in laboratory information management system |url=https://linkinghub.elsevier.com/retrieve/pii/S0169743912001438 |journal=Chemometrics and Intelligent Laboratory Systems |language=en |volume=118 |pages=187–192 |doi=10.1016/j.chemolab.2012.07.001}}</ref> | ||
| Line 142: | Line 136: | ||
====Digital assistants==== | ====Digital assistants==== | ||
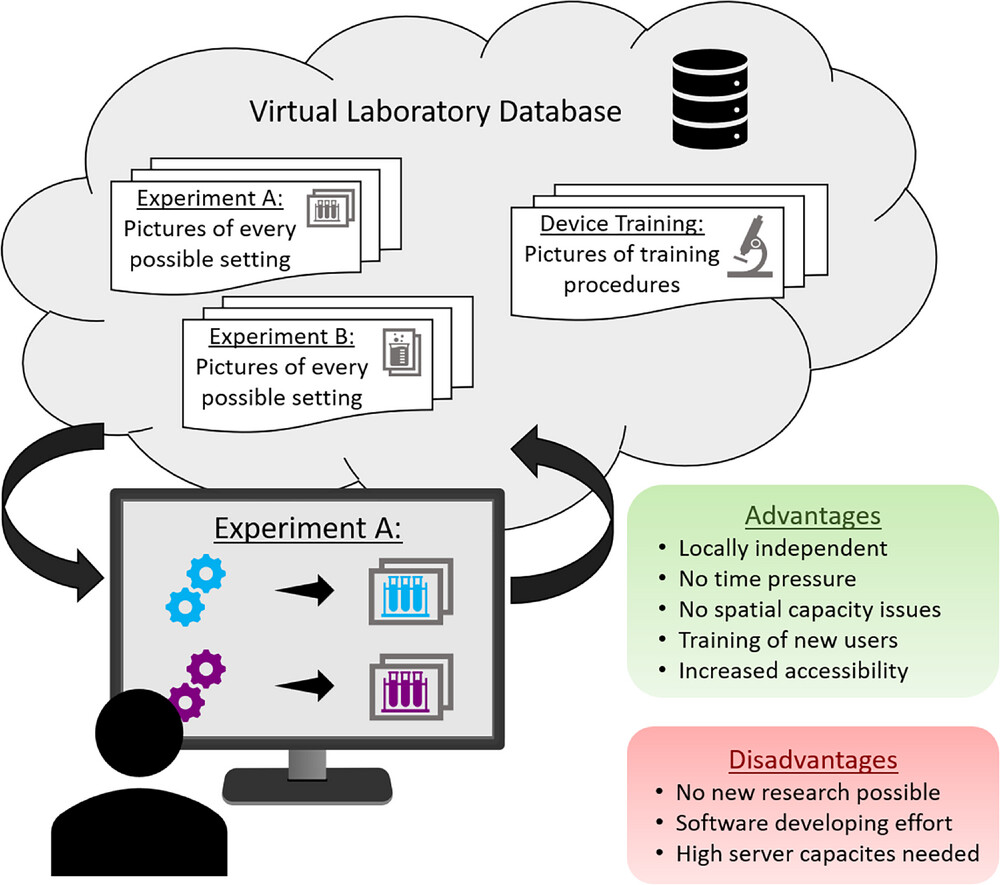
Digital assistants include everything that might help the person working in the laboratory in their digital work, such as workflow scheduling, data gathering, or documenting experiments. The first digital assistant gaining importance in academic laboratories is the ELN. Requirements for the ELN are becoming more and more sophisticated and complex. It is not just about digital documentation of experiments but also about automated data gathering, connectivity to laboratory devices, sharing progress in team projects, as well as improving user-friendliness, data security, research quality. | Digital assistants include everything that might help the person working in the laboratory in their digital work, such as workflow scheduling, data gathering, or documenting experiments. The first digital assistant gaining importance in academic laboratories is the ELN. Requirements for the ELN are becoming more and more sophisticated and complex. It is not just about digital documentation of experiments but also about automated data gathering, connectivity to laboratory devices, sharing progress in team projects, as well as improving user-friendliness, data security, research quality.<ref name=":11" /><ref>{{Cite journal |last=Potthoff |first=Jan |last2=Tremouilhac |first2=Pierre |last3=Hodapp |first3=Patrick |last4=Neumair |first4=Bernhard |last5=Bräse |first5=Stefan |last6=Jung |first6=Nicole |date=2019-03 |title=Procedures for systematic capture and management of analytical data in academia |url=https://linkinghub.elsevier.com/retrieve/pii/S2590134619300039 |journal=Analytica Chimica Acta: X |language=en |volume=1 |pages=100007 |doi=10.1016/j.acax.2019.100007 |pmc=PMC7587027 |pmid=33117974}}</ref> With the ongoing digitalization of academic laboratories, most data are already digitally available and can be linked, copied, or integrated into the ELN. With the increasing availability of ELNs with different sizes and scopes, as well as increasing need for ELNs becoming evident, it is only a matter of time until all laboratories (including academia) will use an ELN instead of an analog laboratory notebook.<ref name=":36">{{Cite journal |last=Dirnagl |first=Ulrich |last2=Przesdzing |first2=Ingo |date=2016-01-04 |title=A pocket guide to electronic laboratory notebooks in the academic life sciences |url=https://f1000research.com/articles/5-2/v1 |journal=F1000Research |language=en |volume=5 |pages=2 |doi=10.12688/f1000research.7628.1 |issn=2046-1402 |pmc=PMC4722687 |pmid=26835004}}</ref><ref>{{Cite journal |last=Voegele |first=Catherine |last2=Bouchereau |first2=Baptiste |last3=Robinot |first3=Nivonirina |last4=McKay |first4=James |last5=Damiecki |first5=Philippe |last6=Alteyrac |first6=Lucile |date=2013-07-01 |title=A universal open-source Electronic Laboratory Notebook |url=https://academic.oup.com/bioinformatics/article/29/13/1710/200295 |journal=Bioinformatics |language=en |volume=29 |issue=13 |pages=1710–1712 |doi=10.1093/bioinformatics/btt253 |issn=1367-4803}}</ref> When switching an entire laboratory or institute to ELN, IT support needs to be considered: data might get stored on servers that need to be maintained; mobile devices, computers, and WIFI needs to be available; and upgrade and backup-support might be needed as well. After switching to an ELN, most users are preferring it over the paper-based version.<ref name=":36" /> When going through the transformation towards a digitalized laboratory, it is important to take concerns raised by future users serious to increase acceptance of new tools and this new way of working in the laboratory. | ||
The next level of digital assistance in bioprocess development laboratories is using a LIMS. The LIMS is where all data are collected, processed, managed, represented, and stored. The ELN is, in most cases, integrated into this management system. Using LIMS will give the laboratory user the advantage to digitally combine various parts of the laboratory such as data gathering, inventory management, workflow scheduling, sample tracking, data mining, device controlling, report creation, information overview, and metadata analysis. | The next level of digital assistance in bioprocess development laboratories is using a LIMS. The LIMS is where all data are collected, processed, managed, represented, and stored. The ELN is, in most cases, integrated into this management system. Using LIMS will give the laboratory user the advantage to digitally combine various parts of the laboratory such as data gathering, inventory management, workflow scheduling, sample tracking, data mining, device controlling, report creation, information overview, and metadata analysis.<ref name=":13" /><ref name=":29" /><ref name=":36" /> Just as ELNs, there are various LIMS that differ in their scope and capabilities.<ref name=":35" /> LIMS is also the basis of starting to enable model-based control of bioprocesses and automating workflows.<ref name=":3" /> Another mean of digital assistance in the lab is the use of smart assistants like Amazon's Alexa. Laboratory workers can ask simple questions about melting points, molecular weight, and isoelectric points, or get answers to simple calculations during a workflow without having to physically interrupt their experiment.<ref>{{Cite web |last=Condliffe, J. |date=03 May 2017 |title=Scientists Are Turning Alexa into an Automated Lab Helper |work=MIT Technology Review |url=https://www.technologyreview.com/2017/05/03/68492/scientists-are-turning-alexa-into-an-automated-lab-helper/}}</ref><ref>{{Cite journal |last=Halford, B. |date=02 May 2017 |title=Meet your new lab assistant |url=https://cen.acs.org/articles/95/i19/Meet-your-new-lab-assistant.html |journal=Chemical & Engineering News |volume=95 |issue=19}}</ref> | ||
====Physical assistants==== | ====Physical assistants==== | ||
Liquid handling stations, autosamplers, and real-time imaging devices are physically helping researchers with laboratory work. In recent years, however, stationary and mobile robots, as well as cobots, are starting to appear sporadically in academic laboratories supporting workers with various physical tasks. | Liquid handling stations, autosamplers, and real-time imaging devices are physically helping researchers with laboratory work. In recent years, however, stationary and mobile robots, as well as cobots, are starting to appear sporadically in academic laboratories supporting workers with various physical tasks.<ref>{{Cite journal |last=Liu |first=Hui |last2=Stoll |first2=Norbert |last3=Junginger |first3=Steffen |last4=Thurow |first4=Kerstin |date=2013-07-01 |title=Mobile Robot for Life Science Automation |url=http://journals.sagepub.com/doi/10.5772/56670 |journal=International Journal of Advanced Robotic Systems |language=en |volume=10 |issue=7 |pages=288 |doi=10.5772/56670 |issn=1729-8814}}</ref> Stationary robots are usually fixed to a place in the laboratory, unable to change their location by themselves, thus being mainly used to execute clear and repetitive workflows. Mounting them on rails gives them added spatial flexibility, increasing their potential application possibilities.<ref name=":4" /> Mobile robots are taking on the task of transporting objects or samples inside the laboratory using their sensors, actuators, and information processing for navigation.<ref name=":4" /><ref name=":34" /> These mobile robots might even be drones in the future that use the air space in laboratories as transportation routes.<ref name=":4" /> They are independent from human cooperation, mostly needing input or commands to start but then work on their own. The development of sensors and also the setup of the whole laboratory or building needs to be considered when using mobile robots. When they transport an object to a different floor, there needs to be some kind of elevator, automatic doors, and no unplanned obstacles in the way.<ref name=":15" /> | ||
Cobots on the other hand are collaborative robots that can be used in cooperation with humans and, due to their light weight design and improvement of safety features, they can also be used close to humans. | Cobots on the other hand are collaborative robots that can be used in cooperation with humans and, due to their light weight design and improvement of safety features, they can also be used close to humans.<ref name=":4" /><ref>{{Cite journal |last=Ghandour, M.; Liu, H.; Stoll, N. et al. |year=2016 |title=A hybrid collision avoidance system for indoor mobile robots based on human-robot interaction |url=https://ieeexplore.ieee.org/document/7827795 |journal=Proceedings of the 17th International Conference on Mechatronics - Mechatronika (ME) |pages=1–7}}</ref> Robots in general are not common in academic laboratories yet, but researchers are starting to see the advantages that robots (stationary and mobile) and cobots offer. They rule out human volatility errors, do not mislabel samples, always transfer the right liquid or the right sample to the intended place, and will always log the right parameters.<ref name=":7" /> Long development times, high maintenance needs, high monetary investments, and even reconstruction of whole laboratory areas are hindering the progress of increasing the amount of physical help in form of robots in academic laboratories. Additionally, in order for robots to be implemented into the academic laboratory, a digital infrastructure with a suited LIMS needs to be implemented first.<ref name=":34" /> However, using robotics in the laboratory is helping to automate workflows or even whole processes, which will be further discussed in the following subsection. | ||
===Automated laboratories=== | ===Automated laboratories=== | ||
A high level of automation is possible for a few laboratories in the field of academic bioprocess development. However, automation and high-throughput is not possible for most experimental settings in research laboratories due to the need of high flexibility and high variety of process workflows. | A high level of automation is possible for a few laboratories in the field of academic bioprocess development. However, automation and high-throughput is not possible for most experimental settings in research laboratories due to the need of high flexibility and high variety of process workflows.<ref name=":4" /> The complexity of academic laboratories is also justified in the high diversity of laboratory staff composition. It is not rare that personnel from more than one research project are sharing laboratories and or devices resulting in a different use of those. More challenges on the way to automating workflows are organizing the setup and ordering system<ref name=":37">{{Cite journal |last=Blow |first=Nathan |date=2008-01 |title=Lab automation: tales along the road to automation |url=https://www.nature.com/articles/nmeth0108-109 |journal=Nature Methods |language=en |volume=5 |issue=1 |pages=109–112 |doi=10.1038/nmeth0108-109 |issn=1548-7091}}</ref>, integrating standalone devices that are already automated by themselves<ref name=":5" />, matching software requirements like legacy technology, proprietary interfaces, and new standard communication protocols (SiLA2, OPC UA).<ref name=":5" /> | ||
To collect as much data as possible for optimization of a bioprocess, Schmid ''et al.'' automated a setup for a DoE for cultivation experiments. | To collect as much data as possible for optimization of a bioprocess, Schmid ''et al.'' automated a setup for a DoE for cultivation experiments.<ref name=":5" /> Using iLAB as the software framework, they integrated a liquid handling station and the DoE software MODDE into their digital infrastructure. The process parameters of cultivation experiments performed by two different bioreactor systems (96-well plate and 10 mL scale) were gathered and analyzed automatically in order to optimize the bioprocess. The integration of the liquid handling station and the standalone software MODDE, which was used to plan the experiments, was realized using SiLA2 and an iLAB plugin. This highly specific automated workflow made it possible to compare data and results from variously scaled experiments due to the focus on data integrity throughout the entire automated process.<ref name=":5" /> | ||
In the laboratory group of Buzz Baum at the University College London, Jennifer Rohn set up an automated microscopy for high-throughput screening of genes. Using an automated liquid handling system, short interfering RNA (siRNA), and automated microscopy for imaging, they are now able to screen genome-wide for genes that play a role in different morphological properties of human cells. After they overcame struggles to set up the workflow in the beginning, considering basic needs like freezer space or inventory control, they also found it challenging to find a good algorithm for analyzing the cell morphology. Using the automated setup saved their group a lot of time and made it possible to exploit a lot more of the available technology. | In the laboratory group of Buzz Baum at the University College London, Jennifer Rohn set up an automated microscopy for high-throughput screening of genes. Using an automated liquid handling system, short interfering RNA (siRNA), and automated microscopy for imaging, they are now able to screen genome-wide for genes that play a role in different morphological properties of human cells. After they overcame struggles to set up the workflow in the beginning, considering basic needs like freezer space or inventory control, they also found it challenging to find a good algorithm for analyzing the cell morphology. Using the automated setup saved their group a lot of time and made it possible to exploit a lot more of the available technology.<ref name=":37" /> | ||
Seidel ''et al.'' developed a comprehensive IT infrastructure for data handling and device integration. | Seidel ''et al.'' developed a comprehensive IT infrastructure for data handling and device integration.<ref name=":13" /> Thus, they were able to automate a bioprocess workflow from online selection of enzymes through ordering gene sequences, cloning, transformation, cultivation, and preproduction to the final scale up to production scale. They focused on using a software that is comprehensive and easy to understand. Human error and workload of laboratory personnel as well as bioprocess developing times are reduced. Even though data storage and sharing is improved using their IT infrastructure, maintenance costs for databases, devices, and interfaces have increased. The whole digital infrastructure is only user-friendly as long as it helps the laboratory personnel in their tasks rather than making it more complicated.<ref name=":13" /> | ||
The potential of fully automated workflows or even fully automated laboratories in academic bioprocess development laboratories is yet to be discovered and will also not be possible everywhere due to the need of high flexibility and complexity. Dividing whole processes into sub-processes, developing a digital twin, using a modular setup, and staying flexible throughout the planning will ultimately help to automate an entire workflow. | The potential of fully automated workflows or even fully automated laboratories in academic bioprocess development laboratories is yet to be discovered and will also not be possible everywhere due to the need of high flexibility and complexity. Dividing whole processes into sub-processes, developing a digital twin, using a modular setup, and staying flexible throughout the planning will ultimately help to automate an entire workflow.<ref name=":3" /><ref name=":4" /><ref name=":13" /><ref>{{Cite journal |last=Schuh |first=Günther |last2=Walendzik |first2=Pia |last3=Luckert |first3=Melanie |last4=Birkmeier |first4=Martin |last5=Weber |first5=Anja |last6=Blum |first6=Matthias |date=2016-11-28 |title=Keine Industrie 4.0 ohne den Digitalen Schatten: Wie Unternehmen die notwendige Datenbasis schaffen |url=https://www.degruyter.com/document/doi/10.3139/104.111613/html |journal=Zeitschrift für wirtschaftlichen Fabrikbetrieb |language=en |volume=111 |issue=11 |pages=745–748 |doi=10.3139/104.111613 |issn=2511-0896}}</ref><ref>{{Cite journal |last=Kuhn |first=Thomas |date=2017-10 |title=Digitaler Zwilling |url=http://link.springer.com/10.1007/s00287-017-1061-2 |journal=Informatik-Spektrum |language=de |volume=40 |issue=5 |pages=440–444 |doi=10.1007/s00287-017-1061-2 |issn=0170-6012}}</ref> | ||
===Artificial intelligence and machine learning=== | ===Artificial intelligence and machine learning=== | ||
[[Artificial intelligence]] (AI) is a general term for the idea of code or algorithms to understand data in a way a human would, or carry out complex tasks based on data. Using AI tools like [[machine learning]] (ML) to analyze collected data can help to generate predictions or knowledge from big datasets efficiently. Besides analyzing simulated experiment data, AI and ML can also be used to solve challenges in everyday laboratory work. Especially when it comes to classification of data sets, protein structure predictions, or image recognition, these increasingly common techniques can prove useful. | [[Artificial intelligence]] (AI) is a general term for the idea of code or algorithms to understand data in a way a human would, or carry out complex tasks based on data. Using AI tools like [[machine learning]] (ML) to analyze collected data can help to generate predictions or knowledge from big datasets efficiently. Besides analyzing simulated experiment data, AI and ML can also be used to solve challenges in everyday laboratory work. Especially when it comes to classification of data sets, protein structure predictions, or image recognition, these increasingly common techniques can prove useful.<ref name=":38">{{Citation |last=Diaz-Flores |first=Ernesto |last2=Meyer |first2=Tim |last3=Giorkallos |first3=Alexis |date=2022 |editor-last=Beutel |editor-first=Sascha |editor2-last=Lenk |editor2-first=Felix |title=Evolution of Artificial Intelligence-Powered Technologies in Biomedical Research and Healthcare |url=https://link.springer.com/10.1007/10_2021_189 |work=Smart Biolabs of the Future |language=en |publisher=Springer International Publishing |place=Cham |volume=182 |pages=23–60 |doi=10.1007/10_2021_189 |isbn=978-3-031-12581-2 |accessdate=2024-04-23}}</ref><ref name=":39">{{Cite journal |last=Nikita |first=Saxena |last2=Thakur |first2=Garima |last3=Jesubalan |first3=Naveen G |last4=Kulkarni |first4=Amey |last5=Yezhuvath |first5=Vinesh B. |last6=Rathore |first6=Anurag S |date=2022-08 |title=AI-ML applications in bioprocessing: ML as an enabler of real time quality prediction in continuous manufacturing of mAbs |url=https://linkinghub.elsevier.com/retrieve/pii/S0098135422002344 |journal=Computers & Chemical Engineering |language=en |volume=164 |pages=107896 |doi=10.1016/j.compchemeng.2022.107896}}</ref> | ||
In 2021, Jumper ''et al.'' published their AI-based program AlphaFold that uses ML to predict the three-dimensional structure of proteins, based on their amino acid sequence, with a high accuracy. | In 2021, Jumper ''et al.'' published their AI-based program AlphaFold that uses ML to predict the three-dimensional structure of proteins, based on their amino acid sequence, with a high accuracy.<ref>{{Cite journal |last=Jumper |first=John |last2=Evans |first2=Richard |last3=Pritzel |first3=Alexander |last4=Green |first4=Tim |last5=Figurnov |first5=Michael |last6=Ronneberger |first6=Olaf |last7=Tunyasuvunakool |first7=Kathryn |last8=Bates |first8=Russ |last9=Žídek |first9=Augustin |last10=Potapenko |first10=Anna |last11=Bridgland |first11=Alex |date=2021-08-26 |title=Highly accurate protein structure prediction with AlphaFold |url=https://www.nature.com/articles/s41586-021-03819-2 |journal=Nature |language=en |volume=596 |issue=7873 |pages=583–589 |doi=10.1038/s41586-021-03819-2 |issn=0028-0836 |pmc=PMC8371605 |pmid=34265844}}</ref> AI-ML applications can help to make process control decisions that will impact the quality of the target product, as Nikita ''et al.'' showed in their work on continuous manufacturing of monoclonal antibodies.<ref name=":39" /> While ML methods helped Volpato ''et al.'' to predict the enzymatic class of a protein based solely on the full amino acid sequence, other works showed the power of ML for classification problems as well. Austerjost ''et al.'' developed a mobile application for an image-based counting algorithm for ''Escherichia coli'' colonies on agar plates. Their idea was to use ML in order to have a more efficient sample throughput with high accuracy.<ref name=":33" /> Biermann ''et al.'' used an algorithm-based image processing for the at-line determination of endospores from microscopic pictures.<ref>{{Cite journal |last=Biermann |first=Riekje |last2=Niemeyer |first2=Laura |last3=Rösner |first3=Laura |last4=Ude |first4=Christian |last5=Lindner |first5=Patrick |last6=Bice |first6=Ismet |last7=Beutel |first7=Sascha |date=2022-03 |title=Facilitated endospore detection for Bacillus spp. through automated algorithm‐based image processing |url=https://onlinelibrary.wiley.com/doi/10.1002/elsc.202100137 |journal=Engineering in Life Sciences |language=en |volume=22 |issue=3-4 |pages=299–307 |doi=10.1002/elsc.202100137 |issn=1618-0240 |pmc=PMC8961035 |pmid=35382541}}</ref> Here, too, the aim was to achieve a faster result using ML. Thus, the presented applications are reducing the time to determine a certain number (''E. coli'' colonies and endospores, respectively), making everyday laboratory work faster and more efficient. | ||
Further and deeper information of ML in the field of bioprocess engineering can be read in the work of Duong-Trung ''et al.'' | Further and deeper information of ML in the field of bioprocess engineering can be read in the work of Duong-Trung ''et al.''<ref>{{Cite journal |last=Duong-Trung |first=Nghia |last2=Born |first2=Stefan |last3=Kim |first3=Jong Woo |last4=Schermeyer |first4=Marie-Therese |last5=Paulick |first5=Katharina |last6=Borisyak |first6=Maxim |last7=Cruz-Bournazou |first7=Mariano Nicolas |last8=Werner |first8=Thorben |last9=Scholz |first9=Randolf |last10=Schmidt-Thieme |first10=Lars |last11=Neubauer |first11=Peter |date=2023-01 |title=When bioprocess engineering meets machine learning: A survey from the perspective of automated bioprocess development |url=https://linkinghub.elsevier.com/retrieve/pii/S1369703X22004338 |journal=Biochemical Engineering Journal |language=en |volume=190 |pages=108764 |doi=10.1016/j.bej.2022.108764}}</ref> As shown in these examples, AI or ML can make laboratory work faster and more efficient. To achieve these results, large amounts of data needed to be generated or harvested from online databases first. This data then needs to be explored, preprocessed, and or annotated manually. Here, education on how to handle big sets of data is beneficial for scientific staff. After data preparation models, neural networks or algorithms need to be developed, trained, and optimized before they can provide the support the laboratory worker is anticipating.<ref name=":38" /> Therefore, to unlock the whole potential of AI or ML in the laboratory, digitalization for automated secure data capture is needed as the first step. | ||
===Simulation of experiments using process models=== | ===Simulation of experiments using process models=== | ||
Digital twins, as previously discussed, have, among other things, potential to gather great amounts of data due to their real-time data monitoring. With advancing digitalization in bioprocess laboratories, the amount of data from experiments is therefore also increasing. Based upon this obtained data, process models can be designed that represent the biotechnological process as a whole to run simulations, with the aim to extract new valuable data. | Digital twins, as previously discussed, have, among other things, potential to gather great amounts of data due to their real-time data monitoring. With advancing digitalization in bioprocess laboratories, the amount of data from experiments is therefore also increasing. Based upon this obtained data, process models can be designed that represent the biotechnological process as a whole to run simulations, with the aim to extract new valuable data.<ref name=":6" /><ref>{{Cite journal |last=Hartmann |first=Fabian S.F. |last2=Udugama |first2=Isuru A. |last3=Seibold |first3=Gerd M. |last4=Sugiyama |first4=Hirokazu |last5=Gernaey |first5=Krist V. |date=2022-11 |title=Digital models in biotechnology: Towards multi-scale integration and implementation |url=https://linkinghub.elsevier.com/retrieve/pii/S0734975022001112 |journal=Biotechnology Advances |language=en |volume=60 |pages=108015 |doi=10.1016/j.biotechadv.2022.108015}}</ref><ref>{{Cite journal |last=Oeing |first=Jonas |last2=Henke |first2=Fabian |last3=Kockmann |first3=Norbert |date=2021-12 |title=Machine Learning Based Suggestions of Separation Units for Process Synthesis in Process Simulation |url=https://onlinelibrary.wiley.com/doi/10.1002/cite.202100082 |journal=Chemie Ingenieur Technik |language=en |volume=93 |issue=12 |pages=1930–1936 |doi=10.1002/cite.202100082 |issn=0009-286X}}</ref> Modeling experiments and simulating their respective outcomes using parameter adjustment is unlocking new potential to get information without having to do all the actual work inside the laboratory.<ref name=":38" /> After a process model has been developed, it is important to verify its accuracy either through experimental validation experiments or through expert knowledge.<ref name=":3" /> | ||
Zobel-Roos ''et al.'' used the data of only a small amount of experiments for a process model to evaluate the outcome of different continuous chromatography setups. These small experiments were done to estimate the parameters for the process model. The results determined by the model were then experimentally verified in the laboratory. Simulation and tracer experiments showed a deviation as small as 0.8%. | Zobel-Roos ''et al.'' used the data of only a small amount of experiments for a process model to evaluate the outcome of different continuous chromatography setups. These small experiments were done to estimate the parameters for the process model. The results determined by the model were then experimentally verified in the laboratory. Simulation and tracer experiments showed a deviation as small as 0.8%.<ref>{{Cite journal |last=Zobel-Roos |first=Steffen |last2=Stein |first2=Dominik |last3=Strube |first3=Jochen |date=2018-03-02 |title=Evaluation of Continuous Membrane Chromatography Concepts with an Enhanced Process Simulation Approach |url=https://www.mdpi.com/2073-4468/7/1/13 |journal=Antibodies |language=en |volume=7 |issue=1 |pages=13 |doi=10.3390/antib7010013 |issn=2073-4468 |pmc=PMC6698847 |pmid=31544865}}</ref> Ladd Effio ''et al.'' showed successful process modeling approaches for the downstream part of a bioprocess optimizing anion-exchange [[chromatography]]. After experimentally determining adsorption isotherms and mass transfer parameters they were subsequently able to use their simulation-based model to identify optimal elution conditions.<ref>{{Cite journal |last=Ladd Effio |first=Christopher |last2=Hahn |first2=Tobias |last3=Seiler |first3=Julia |last4=Oelmeier |first4=Stefan A. |last5=Asen |first5=Iris |last6=Silberer |first6=Christine |last7=Villain |first7=Louis |last8=Hubbuch |first8=Jürgen |date=2016-01 |title=Modeling and simulation of anion-exchange membrane chromatography for purification of Sf9 insect cell-derived virus-like particles |url=https://linkinghub.elsevier.com/retrieve/pii/S0021967315017562 |journal=Journal of Chromatography A |language=en |volume=1429 |pages=142–154 |doi=10.1016/j.chroma.2015.12.006}}</ref> Boi ''et al.'' were able to develop a model of affinity membrane chromatography to help with the prediction of membrane performance in bioprocess design.<ref>{{Cite journal |last=Boi |first=Cristiana |last2=Dimartino |first2=Simone |last3=Sarti |first3=Giulio C. |date=2007-08 |title=Modelling and simulation of affinity membrane adsorption |url=https://linkinghub.elsevier.com/retrieve/pii/S0021967307002658 |journal=Journal of Chromatography A |language=en |volume=1162 |issue=1 |pages=24–33 |doi=10.1016/j.chroma.2007.02.008}}</ref> | ||
Cell type, physiology, metabolism, and more factors are substantially increasing the complexity of bioprocesses when whole cells are involved. While representing a reliable mathematical model of a whole cell is still seemingly impossible, model-based methods are able to make predictions. These predictions cannot be fully trusted and have to be verified with experiments in the laboratory. However, designing bioprocesses by analyzing data with ML methods and simulations can help to optimize experimental setups that can subsequently be transferred to the laboratory. | Cell type, physiology, metabolism, and more factors are substantially increasing the complexity of bioprocesses when whole cells are involved. While representing a reliable mathematical model of a whole cell is still seemingly impossible, model-based methods are able to make predictions. These predictions cannot be fully trusted and have to be verified with experiments in the laboratory. However, designing bioprocesses by analyzing data with ML methods and simulations can help to optimize experimental setups that can subsequently be transferred to the laboratory.<ref name=":1" /><ref name=":3" /> | ||
===Virtual laboratories=== | ===Virtual laboratories=== | ||
Besides digitalizing the physical laboratory, there is another approach that is particularly practical for educational purposes: virtual laboratories. The concept of these laboratories is that results of experiments are being photographed or otherwise documented and stored with their respective settings. These pictures and information are then uploaded to the virtual laboratory (online database). As depicted in Figure 5, users can then access this virtual laboratory through, for example, a graphical web-service, alternating the settings of the virtual experimental setup and receiving the associated picture or outcome. | Besides digitalizing the physical laboratory, there is another approach that is particularly practical for educational purposes: virtual laboratories. The concept of these laboratories is that results of experiments are being photographed or otherwise documented and stored with their respective settings. These pictures and information are then uploaded to the virtual laboratory (online database). As depicted in Figure 5, users can then access this virtual laboratory through, for example, a graphical web-service, alternating the settings of the virtual experimental setup and receiving the associated picture or outcome.<ref>{{Cite journal |last=Torre |first=Luis de la |last2=Neustock |first2=Lars Thorben |last3=Herring |first3=George K. |last4=Chacon |first4=Jesus |last5=Clemente |first5=Felix J. Garcia |last6=Hesselink |first6=Lambertus |date=2020-12 |title=Automatic Generation and Easy Deployment of Digitized Laboratories |url=https://ieeexplore.ieee.org/document/9018197/ |journal=IEEE Transactions on Industrial Informatics |volume=16 |issue=12 |pages=7328–7337 |doi=10.1109/TII.2020.2977113 |issn=1551-3203}}</ref> This tool proves to be useful for a deeper understanding of complex device equipment. A study found that students who completed an online device training in a virtual laboratory before conducting the experiment in the physical lab needed less guidance and were able to spend more time for the actual experiment rather than getting familiar with the complex equipment. Having learned the basic equipment knowledge online, these students were better prepared and more confident than those who did not use the virtual laboratory beforehand.<ref>{{Cite journal |last=Hatherly |first=P A |last2=Jordan |first2=S E |last3=Cayless |first3=A |date=2009-07-01 |title=Interactive screen experiments—innovative virtual laboratories for distance learners |url=https://iopscience.iop.org/article/10.1088/0143-0807/30/4/008 |journal=European Journal of Physics |volume=30 |issue=4 |pages=751–762 |doi=10.1088/0143-0807/30/4/008 |issn=0143-0807}}</ref><ref name=":40">{{Cite journal |last=Jones |first=Nicola |date=2018-10 |title=Simulated labs are booming |url=https://www.nature.com/articles/d41586-018-06831-1 |journal=Nature |language=en |volume=562 |issue=7725 |pages=S5–S7 |doi=10.1038/d41586-018-06831-1 |issn=0028-0836}}</ref> When students use virtual instead of traditional laboratories for educational purposes, there is no difference in the effectiveness of content knowledge.<ref>{{Cite journal |last=Faulconer |first=Emily Kaye |last2=Gruss |first2=Amy B |date=2018-05-01 |title=A Review to Weigh the Pros and Cons of Online, Remote, and Distance Science Laboratory Experiences |url=http://www.irrodl.org/index.php/irrodl/article/view/3386 |journal=The International Review of Research in Open and Distributed Learning |volume=19 |issue=2 |doi=10.19173/irrodl.v19i2.3386 |issn=1492-3831}}</ref> The easy and automated generation of virtual laboratories is well described in the work of De La Torre ''et al.''<ref name=":41">{{Cite journal |last=Torre |first=Luis de la |last2=Neustock |first2=Lars Thorben |last3=Herring |first3=George K. |last4=Chacon |first4=Jesus |last5=Clemente |first5=Felix J. Garcia |last6=Hesselink |first6=Lambertus |date=2020-12 |title=Automatic Generation and Easy Deployment of Digitized Laboratories |url=https://ieeexplore.ieee.org/document/9018197/ |journal=IEEE Transactions on Industrial Informatics |volume=16 |issue=12 |pages=7328–7337 |doi=10.1109/TII.2020.2977113 |issn=1551-3203}}</ref> Even if this digitalization strategy has no impact for academic laboratories to generate new research data, this can well be used for training or educational purposes.<ref name=":41" /> | ||
| Line 191: | Line 185: | ||
|} | |} | ||
The use of virtual reality (VR) is another way to interpret the term virtual laboratories. For better understanding, the terms "VR" and "augmented reality" (AR) are shortly explained by Nakhle ''et al.'' | The use of virtual reality (VR) is another way to interpret the term virtual laboratories. For better understanding, the terms "VR" and "augmented reality" (AR) are shortly explained by Nakhle ''et al.''<ref name=":42">{{Cite journal |last=Nakhle |first=Farid |last2=Harfouche |first2=Antoine L. |date=2023-01 |title=Extended reality gives digital agricultural biotechnology a new dimension |url=https://linkinghub.elsevier.com/retrieve/pii/S0167779922002396 |journal=Trends in Biotechnology |language=en |volume=41 |issue=1 |pages=1–5 |doi=10.1016/j.tibtech.2022.09.005}}</ref>: | ||
* Virtual reality: “Immerses users into interactive and explorative virtual environments based on 3D representation and simulation of the real world and objects within it.” Requirements: VR headset, PC. | *Virtual reality: “Immerses users into interactive and explorative virtual environments based on 3D representation and simulation of the real world and objects within it.” Requirements: VR headset, PC.<ref name=":42" /> | ||
* Augmented reality: “Superimposes valuable visual content and information over a live view of the real world to provide users an enriched version of reality, allowing them to interact with digital objects in real-time.” Requirements: Smartphone & tablet, AR glasses. | *Augmented reality: “Superimposes valuable visual content and information over a live view of the real world to provide users an enriched version of reality, allowing them to interact with digital objects in real-time.” Requirements: Smartphone & tablet, AR glasses.<ref name=":42" /> | ||
Harfouche ''et al.'' showed that when studying biotechnology, bioethics can be taught via VR, making a virtual classroom experience possible for students. | Harfouche ''et al.'' showed that when studying biotechnology, bioethics can be taught via VR, making a virtual classroom experience possible for students.<ref name=":43">{{Cite journal |last=Harfouche |first=Antoine L. |last2=Nakhle |first2=Farid |date=2020-11 |title=Creating Bioethics Distance Learning Through Virtual Reality |url=https://linkinghub.elsevier.com/retrieve/pii/S0167779920301268 |journal=Trends in Biotechnology |language=en |volume=38 |issue=11 |pages=1187–1192 |doi=10.1016/j.tibtech.2020.05.005 |pmc=PMC7211661 |pmid=32446631}}</ref> Another example for education via VR is the Arizona State University, which is offering a biology degree solely based on VR laboratory work.<ref name=":40" /><ref>{{Cite web |title=Online Bachelor of Science in Biological Sciences |work=ASU Online |url=https://asuonline.asu.edu/online-degree-programs/undergraduate/biological-sciences-bs/ |publisher=Arizona State University}}</ref> Advantages of these VR classrooms or laboratories include the student's local independence, no exposure to toxic chemicals, no spatial limitations, the possibility to slow down very fast reactions for visualization, and no safety concerns when learning about, for example, explosives or toxins. Thus, the learning experience using VR will be enhanced in a cost and time effective manner. Additionally, students will interactively be taught with reduced human bias regarding gender, age, ethnicity, and race.<ref name=":40" /><ref name=":42" /><ref name=":43" /> | ||
VR can be a helpful tool for scientists to visualize 3D images to grasp cell's morphological features with confocal microscopy. ConfocalVR is a VR application that can be installed on consumer-grade VR systems to visualize microscopic image stacks into 3D images in the virtual world. The user can then interact with the digital object and examine its properties in a way that would not be possible with 2D imaging on a computer screen. | VR can be a helpful tool for scientists to visualize 3D images to grasp cell's morphological features with confocal microscopy. ConfocalVR is a VR application that can be installed on consumer-grade VR systems to visualize microscopic image stacks into 3D images in the virtual world. The user can then interact with the digital object and examine its properties in a way that would not be possible with 2D imaging on a computer screen.<ref>{{Cite journal |last=Stefani |first=Caroline |last2=Lacy-Hulbert |first2=Adam |last3=Skillman |first3=Thomas |date=2018-10 |title=ConfocalVR: Immersive Visualization for Confocal Microscopy |url=https://linkinghub.elsevier.com/retrieve/pii/S0022283618306648 |journal=Journal of Molecular Biology |language=en |volume=430 |issue=21 |pages=4028–4035 |doi=10.1016/j.jmb.2018.06.035 |pmc=PMC6805139 |pmid=29949752}}</ref> | ||
Device training and or maintenance can be improved by using AR. Device experts do not have to travel to laboratories but can guide the scientists through maintenance procedures with instructions while being able to see what the scientists who wears AR glasses sees. Instructions and information can be displayed in the user's view for guidance. This way requesting assistance from local independent experts, teachers, or colleagues is not more than a call away linking more knowledge together. | Device training and or maintenance can be improved by using AR. Device experts do not have to travel to laboratories but can guide the scientists through maintenance procedures with instructions while being able to see what the scientists who wears AR glasses sees. Instructions and information can be displayed in the user's view for guidance. This way requesting assistance from local independent experts, teachers, or colleagues is not more than a call away linking more knowledge together.<ref>{{Cite journal |last=Nee |first=A.Y.C. |last2=Ong |first2=S.K. |last3=Chryssolouris |first3=G. |last4=Mourtzis |first4=D. |date=2012 |title=Augmented reality applications in design and manufacturing |url=https://linkinghub.elsevier.com/retrieve/pii/S0007850612002090 |journal=CIRP Annals |language=en |volume=61 |issue=2 |pages=657–679 |doi=10.1016/j.cirp.2012.05.010}}</ref><ref>{{Cite journal |last=Mourtzis |first=Dimitris |last2=Siatras |first2=Vasileios |last3=Angelopoulos |first3=John |date=2020-03-08 |title=Real-Time Remote Maintenance Support Based on Augmented Reality (AR) |url=https://www.mdpi.com/2076-3417/10/5/1855 |journal=Applied Sciences |language=en |volume=10 |issue=5 |pages=1855 |doi=10.3390/app10051855 |issn=2076-3417}}</ref><ref>{{Cite journal |last=Lamberti |first=Fabrizio |last2=Manuri |first2=Federico |last3=Sanna |first3=Andrea |last4=Paravati |first4=Gianluca |last5=Pezzolla |first5=Pietro |last6=Montuschi |first6=Paolo |date=2014-12 |title=Challenges, Opportunities, and Future Trends of Emerging Techniques for Augmented Reality-Based Maintenance |url=http://ieeexplore.ieee.org/document/7004838/ |journal=IEEE Transactions on Emerging Topics in Computing |volume=2 |issue=4 |pages=411–421 |doi=10.1109/TETC.2014.2368833 |issn=2168-6750}}</ref> | ||
==Conclusions== | ==Conclusions== | ||
| Line 212: | Line 206: | ||
==Abbreviations, acronyms, and initialisms== | ==Abbreviations, acronyms, and initialisms== | ||
* '''AI''': artificial intelligence | |||
* '''AR''': augmented reality | *'''AI''': artificial intelligence | ||
* '''CLI''': command line interface | *'''AR''': augmented reality | ||
* '''DoE''': design of experiments | *'''CLI''': command line interface | ||
* '''ELN''': electronic laboratory notebook | *'''DoE''': design of experiments | ||
* '''FAIR''': findable, accessible, interoperable, reusable | *'''ELN''': electronic laboratory notebook | ||
* '''GUI''': graphical user interface | *'''FAIR''': findable, accessible, interoperable, reusable | ||
* '''IoT''': internet of things | *'''GUI''': graphical user interface | ||
* '''LADS''': Laboratory and Analytical Device Standard | *'''IoT''': internet of things | ||
* '''LIMS''': laboratory information management system | *'''LADS''': Laboratory and Analytical Device Standard | ||
* '''ML''': machine learning | *'''LIMS''': laboratory information management system | ||
* '''NUI''': natural user interface | *'''ML''': machine learning | ||
* '''OPC UA''': open platform communications unified architecture | *'''NUI''': natural user interface | ||
* '''SiLA2''': Standardization in Lab Automation 2 | *'''OPC UA''': open platform communications unified architecture | ||
* '''VR''': virtual reality | *'''SiLA2''': Standardization in Lab Automation 2 | ||
* '''VUI''': voice user interface | *'''VR''': virtual reality | ||
*'''VUI''': voice user interface | |||
==Acknowledgements== | ==Acknowledgements== | ||
Revision as of 22:57, 23 April 2024
| Full article title | Digitalization concepts in academic bioprocess development |
|---|---|
| Journal | Engineering in Life Sciences |
| Author(s) | Habich, Tessa; Beutel, Sascha |
| Author affiliation(s) | Leibniz University Hannover |
| Primary contact | Email: beutel at iftc dot uni dash hannover dot de |
| Year published | 2024 |
| Volume and issue | 24(4) |
| Article # | 2300238 |
| DOI | 10.1002/elsc.202300238 |
| ISSN | 1618-2863 |
| Distribution license | Creative Commons Attribution-NonCommercial 4.0 International |
| Website | https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/elsc.202300238 |
| Download | https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/pdfdirect/10.1002/elsc.202300238 (PDF) |
Abstract
Digitalization with integrated devices, digital and physical assistants, automation, and simulation is setting a new direction for laboratory work. Even with complex research workflows, high staff turnover, and a limited budget, some laboratories have already shown that digitalization is indeed possible. However, academic bioprocess laboratories often struggle to follow the trend of digitalization. Due to their diverse research circumstances, high variety of team composition, goals, and limitations, the concepts are substantially different. Here, we will provide an overview on different aspects of digitalization and describe how academic research laboratories successfully digitalized their working environment. The key aspect is the collaboration and communication between IT-experts and scientific staff. The developed digital infrastructure is only useful if it supports the laboratory worker and does not complicate their work. Thereby, laboratory researchers have to collaborate closely with IT experts in order for a well-developed and maintainable digitalization concept that fits their individual needs and level of complexity to emerge. This review may serve as a starting point or a collection of ideas for the transformation toward a digitalized bioprocess laboratory.
Keywords: academic laboratories, automation, bioprocess, digitalization, FAIR data, Laboratory and Analytical Device Standard (LADS), Standardization in Lab Automation 2 (SiLA2)
Introduction
In contrast to industry, academic research laboratories require more flexibility than production lines. Besides the need for flexibility, loss of knowledge due to high staff turnover in universities is another challenge that makes full laboratory digitalization hard to achieve.[1][2][3][4] "Digitalization" (what we'll be addressing) refers to an entire process, workflow, or laboratory infrastructure, whereas "digitization" refers only to the procedure of converting something analog to a digital format (e.g., digitizing a standard operating procedure [SOP] from a piece of paper to a digital file).[5][6] Working in digital laboratories has the potential for error reduction, prevention of data loss, improved data integrity, faster workflow development times, possible reduction of chemical and material use, and higher sample throughput, leading to modern, transparent, and reproducible research and biomanufacturing.[7][8][9][10][11][12][13][14]
Developing a digitalization strategy for an academic bioprocess laboratory is an interdisciplinary task. Laboratory workers have to work closely with IT experts in order to achieve a digital infrastructure that can be maintained and is supporting their laboratory work rather than hindering it.[5][7][9][15][16] This kind of collaboration is rarely found in academic bioprocess laboratories, so even the smallest digitalization task can be a major challenge. Hardware vendors selling their devices with proprietary software and restrictive access and information about their interfaces is making digitalization even more difficult.[1][7][17] Therefore, academic researchers working in the field of digitalization are developing concepts that are fitting their individual needs at the time. This is why there are manifold digitalization concepts throughout academic laboratories.
This review will present different digitalization concepts throughout academic bioprocess development laboratories. Before looking at different individual concepts in more detail, general aspects like FAIR data (data that is readily findable, accessible, interoperable, and reusable), standard communication protocols, digital twins, and interaction with laboratory devices will be addressed.
Basic concepts of digitalization
FAIR data
With the progressing digitalization of laboratories, the generated amount of data is steadily increasing. Therefore, good data management systems will become inevitable in bioprocess development laboratories. The first step towards good data management is storing the data and metadata according to the FAIR data principles.[18] Both humans and machines should be able to find the data with metadata and a clear unique identifier. The data should be digitally accessible for the user with the appropriate tool. It should be noted that accessible in this context does not mean that the data are "open" or "free" but that transparency around the used data concerning its availability is given.[19][20] Interoperable data means data are presented using vocabulary and data encoding that follows the FAIR data principles and can be read by machines. In order for data to be reusable they need to be rich in metadata and descriptive documentation on the circumstances in which the data were generated. Rich metadata should ideally describe the data in a meaningful way, including the setup and context of the experiment, technical setting, and information about the provenance.[8][18][21][22][23]
Most research data are currently stored locally and not organized in a standardized way. Storing data according to FAIR data principles allows them to be read by machines and not just the human laboratory worker who generated them. Other advantages of following the FAIR data principles, as well as publishing research data, is that this data will stay available over time. Available or even openly online shared data, including metadata, can help to determine if the publication is of high quality. It can be reproduced, reanalyzed, used for new analyses, or even compared to or combined with other data promoting a deeper understanding of the topic or perhaps generating new knowledge.[12][21][24]
Before computers were used on a large scale in laboratories, results of experiments were handwritten into paper notebooks. This is still happening in many cases in today's bioprocess development laboratories, although the use of electronic laboratory notebooks (ELNs) is emerging quickly.[25] Most devices are equipped with a USB, RS232, or RS485 port, thus providing network connectivity. But the absence of a physical network necessitates the usage of, for example, flash drives for data transport. This increases the danger of manipulation or loss of data.[9] Writing down data manually and then digitizing it later on a computer used for data storage and analysis is also not a good solution. The analog handwritten data is prone to error, not good to search through, only accessible from the analog laboratory notebook, and has limited reusability, in contrast to the FAIR data principles.[9][26] After it has been digitized without human errors, it has all the advantages of digital data. In order to make data gathering faster, simpler, and more accurate, a complete transition to automated data capture has to be made.[26] This includes data flow from a whole bioprocess, including sampling, sample preparation, measurement, and data analysis.[8] When transitioning from analog to digital data, it is important to take the laboratory users needs into consideration. While aiming to automatically store data following FAIR data principles, the end user who is analyzing it still needs to be able to access and work with it.
Standard communication protocols
While a lot of laboratory device vendors still offer their own proprietary software for device control[17][27], researchers are agreeing that the future of digitalization in the laboratory lies in the common use of standardized device communication protocols.[7][9][17][26][28][29][30][31][32][33][34] To connect all laboratory devices to one digital infrastructure, it is aspirational for all devices to use the same standard communication protocol to achieve a plug-and-play environment in bioprocess laboratories.[26] Device integration is faster, easier, and better to maintain when vendor-independent communication protocols are more common.[35]
While offering device control with standard communication protocols might be costly for vendors in the beginning due to initial software development, it offers advantages as well. Using a communication standard as an instrument vendor will reduce the complexity of their software documentation since the basics have already been well documented. Thus, a detailed documentation of manufacturer-specific proprietary device drivers will become redundant. This simplification of software development can then reduce costs for device vendors. When using standard protocols for device communication, instrument vendors are contributing to the management of data according to the FAIR data principles, leaving them to focus on new innovations and features in their devices. Customers can then choose devices based upon those innovations instead of technical practicalities. Additionally, more companies and laboratories have the ability to integrate devices with standard communication protocols into their digital infrastructures because they are not limited by interface or platform incompatibilities. This will then lead to an increased purchase of consumables for those devices.[36][37][38][39][40] Agreeing on physical standards like the microtiter plate was an advantage for everyone in the end.[30] Using standardized tools and parts in the production process is decreasing costs and complexity due to purchasing larger quantities of the same part rather than needing multiple specific parts. The earlier standards are getting adopted and implemented in the working process the more cost efficient it will be. Manufacturers are also challenged to differentiate their product from other manufacturers in other areas than compatibility. This leads to a better competition and an overall increase in developing new technologies rather than sticking to proprietary solutions.[30][41] For consumers on the other hand, standardization will help to easily maintain devices even if the manufacturer no longer exists. Standards need to be flexible enough to go beyond their intended purpose or otherwise it will limit their use.
Before implementing a communication standard, a holistic plan needs to be made from scientists in cooperation with IT experts. Laboratory work needs to be simplified, and the scientist needs to be supported through standard device connectivity. Users need to have a clear benefit from using a standard, otherwise digitalization efforts will not be rewarded by regular use. As long as the use of proprietary device control software is more convenient, the use of standard communication protocols is ineffective. These benefits can, for example, be achieved by automating parts of the workflow that would otherwise result in tedious manual work, like data acquisition and central data storage following FAIR data principles. Other means to support the user in the laboratory are semi-automated SOPs, where device parameters are already preset after digitally selecting the required workflow. Using a standard protocol for device communication can realize these aspects when all devices are connected by a central device control server, as demonstrated by Porr et al.[35] and Rachuri et al.[42] After successfully choosing and implementing standard communication, it is important to keep the infrastructure open to new device additions and upgrades without being a burden on laboratory workers. Two standard communication protocols are currently coexisting in bioprocess development laboratories: Standardization in Lab Automation 2 (SiLA2) and the Laboratory and Analytical Device Standard (LADS), based on Open Platform Communications Unified Architecture (OPC UA).[17][36][38][40][43][44]
SiLA2
“SiLA's mission is to establish international standards which create open connectivity in lab automation, to enable lab digitalization.”[38] This open-source communication standard is based on a client-server infrastructure where every laboratory device is a SiLA2 server that offers a set of features. SiLA2 servers advertise themselves on the network and are discovered by SiLA2 clients like a laboratory information management system (LIMS) or the SiLA2 Manager developed by Bromig et al.[36][40][42] The clients recognize the offered features by the servers and can use or call them. Real-time observation of parameters is possible through the subscription to observable properties. For integration of legacy devices, Porr et al. developed a gateway module to enable SiLA communication.[45] On the software side, the gateway module has the advantage of clearly separating the tasks while still using a standard protocol. This plays a major role in scalability and maintainability. SiLA2 has been implemented in different frameworks using Java, C#, Python, or C++ for fast and easy integration of laboratory devices.[42][45][46]
LADS based on OPC UA
OPC UA is a standard in multiple industries for data exchange between devices independent from the device vendor.[44] Just like SiLA2, OPC UA has a client-server infrastructure. OPC clients can be connected to any OPC server that is able to collect and further process provided data from the client. The previous version, OPC Classic, was limited to the operating system Microsoft Windows; however, OPC UA is platform-independent. OPC was used as an established standard in industry even before the need for a standard communication protocol for laboratory equipment manifested itself.[44] The problem of transferring this digital communication standard from industry to bioprocess development research laboratories is the added flexibility and heterogeneity of devices that make those laboratories a lot more complex than industrial production processes when looking at the digitalization part.[43] This is why a working group from the industrial association Spectaris developed the communication protocol LADS based on OPC UA.[17][47] “The goal of LADS is to create a manufacturer-independent, open standard for analytical and laboratory equipment.”[48] OPC UA provides a way to integrate legacy devices into the digital infrastructure by providing wrappers for existing software.[49] Implementing OPC UA is possible in different programming languages such as Java, Python, C++, or C#.[49] OPC UA is not just enabling digital connectivity inside the laboratory but also connecting research laboratories to industrial infrastructures due to OPC UA already being established in industrial environments.[17]
Digital twins
Through the development of Industry 4.0 and progressing internet of things (IoT) connectivity, the number of digital twins is increasing also in the environment of bioprocess engineering laboratories. IoT in this context describes the idea to connect physical objects like laboratory devices to a network or the internet to ensure status and information processing, as well as the ability to communicate and send and receive data.[28][50][51][52][53] This omnipresent keyword "digital twin" is described by Fuller et al. as the “effortless integration of data between a physical and virtual machine in either direction.”[54] This concept is an invaluable advancement in bioprocess development, whereas a differentiation between "digital models," "digital shadows," and "digital twins" is important. The difference between these, as depicted in Figure 1, are the manual and automated data flows between the physical and digital object. All three concepts have a physical and digital object that are connected by data flow. Within the digital model, the data in transported manually between the physical and digital object. The digital shadow has automated data flow from the physical to the digital object, but manual data flow from the digital to the physical object. A digital twin, however, has automated data flow in both directions. Thus, only when using a digital twin, changes in the physical object will result in changes in the digital object and vice versa.[7][54][55]
|
Major challenges in the process of implementing digital twins in the bioprocess laboratory are data security and privacy concerns, device connectivity, trust issues regarding the use of technology, and development of a laboratory-wide digital infrastructure, including data storage space. Difficult to predict and complex processes especially in the biological laboratory complicate the development of digital twins as well. The predominant benefits on the other hand include automated data (and metadata) generation, processing, and analyzing of those as well as data traceability. All this is contributing to the aforementioned FAIR data principles. Laboratories that use digital tracking of consumables in their digital twins are able to automate their ordering system making manual inventory obsolete. Therefore, digital twins are the key technology to a new and transformed laboratory work, with the potential to automated knowledge generation.[7][55]
Interaction with laboratory devices
Besides using standardized device communication protocols, full digitalization cannot be achieved without considering the end user. Digital solutions assisting the laboratory worker but also interacting with devices should therefore be straightforward and convenient to use. The device interactions with laboratory equipment in bioprocess development laboratories has changed over the last years, making it more efficient and intuitive with every additional solution, providing more interaction possibilities.[56]
The use of laboratory devices itself is supposed to not interfere with the experiment, slow it down, or complicate it. Consequently, device operation should be as intuitive as possible. There are three different interactions levels for device control. Starting at the lowest level, there is abstract interaction, using command line interfaces (CLI), followed by the next level, the indirect interaction, using graphical user interfaces (GUI). The last and most intuitive level of device interaction is the direct interaction using natural user interfaces (NUI).
CLIs have the advantage of keyboard input from the user and are easy to integrate or combine with other tools like scripts. However, using CLIs as the interaction medium within the laboratory environment requires basic knowledge of command line logic and scripting. This skill cannot be expected from everyone working in the laboratory due to their different educational backgrounds. One approach to accelerate the transformation towards digitalized laboratories is the addition of basic IT classes (e.g., for basic scripting knowledge) to biotechnological education. However, CLIs as the only device interface are not a suitable solution for general and intuitive device interaction. Yet additional CLI device interaction may prove useful for administrative purposes or for integrating supplementary software.[56]
The next level of interaction with laboratory devices is the indirect interaction using GUIs. This has the advantage that no programming or scripting knowledge is needed for device interaction. Thus, all people in the laboratory are able to operate devices equipped with a GUI. Additionally, more information can be displayed for the user on the screen while mouse and keyboard input is enabled. A lot of devices have a vendor-specific and often proprietary control software. This often runs on a desktop PC positioned next to the device inside the laboratory. Modern devices, however, do not only have the possibility of local interaction but also offer remote interaction via network connection. Those GUIs are then deployed as touch-based network-like web interfaces, also allowing the usage of handheld devices as the end device. These handheld devices like smartphones, tablets, or touch beamers have the advantage that people are used to operating touch displays from their personal life making device interaction even more intuitive. Additionally, the use of mobile applications on smartphones and tablets is enabled.[57][58][59]
The use of touch-based interaction media is introducing the next level of device interaction: NUI. Besides touch-based interaction media, NUIs also include voice user interfaces (VUI) and interaction via gestures and motions. Artificial input like keyboard and mouse input becomes obsolete. This direct interaction with laboratory devices is leading to a simplified and even more intuitive way of working in the digitalized laboratory environment. Even though NUIs have a lot of advantages (Figure 2) they are not undisputed due to security concerns or voice recognition issues.[60][61] Austerjost et al. showed in their work that VUIs can enable hands-free device control, which can be particularly advantageous in microbiology laboratories regarding contamination risks.[34] The topic of human-device interaction is described in further detail in the work of Söldner et al.[53]
|
Digitalization strategies
The way digitalization in laboratories looks can be very different everywhere. Bioprocess development is still relying on experienced workers doing experiments in the laboratory and analyzing data afterwards. Digitalization with integrating devices, digital and physical assistants, automation, and simulation is setting a new direction for laboratory work.[2][62] One digitalization strategy alone will not be able to meet all needs of the various laboratories that exists in the academic bioprocess development field.[30] In the following subsections, different approaches of digitalization with their respective applications are presented and discussed.
Digital integration of laboratory devices
The first step to digitalize any laboratory is to connect devices to a digital infrastructure. This connection can look different for each laboratory, depending on their individual needs. Due to high topic variance in some academic research laboratories, the devices are usually waiting to be used by a researcher in their required manual way.[62] Devices in these laboratories are described by Porr et al. as a “craftsman's toolbox.”[45] Integrating them into a highly automated workflow might hinder research and flexibility, which is why there are different approaches to hardware integration in academic research laboratories. Laboratory devices can be divided into three categories with different levels of technical prerequisites. In order to connect or control any device with a digital infrastructure, a central device control server needs to be developed first. The SiLA2 manager developed by Bromig et al.[42] or the DeviceLayer from Porr et al.[35] are examples of this. The connection to different laboratory devices will then look different due to their category. While legacy laboratory devices do not have internet access connection points but rather a USB or other serial connection ports (like RS232), they first need to be connected to a physical device that bridges the connection to the network as well as to the central control server and enables standard communication. This can be achieved using a gateway module. Laboratory devices with network access (WLAN or Ethernet) do not need such a physical device in order to be connected using a standard device communication protocol. A simple digital component like a device controller is sufficient to integrate that device into the digital infrastructure. Laboratory devices that come with a standard driver like SiLA2 or OPC UA can be controlled directly from the central control server.[35] Figure 3 shows this concept with SiLA2 as the example for the standard device communication protocol.
|
For the integration of legacy laboratory devices, Porr et al. developed a gateway module to enable SiLA-based communication. This gateway module is based on an embedded computer that runs a Linux image. The legacy device is connected to this module via USB or RS232 and a SiLA2 server on the other side that can control the device hosted on the gateway module. Thus, communication using the SiLA communication protocol is enabled. In their work, they showed how to use this connection device to integrate a magnetic stirrer into the digital infrastructure.[45] Bromig et al. also connected their laboratory devices using the SiLA2 communication protocol in their work. They developed a SiLA2 Manager that is able to discover, connect to and control SiLA2 drivers generated with SiLA2 Python. Using this software framework, they connected various laboratory devices needed for their use case cultivation workflow with intermittent glucose feeding.[42]
Schmid et al. used the iLAB software framework (meanwhile known as zenLAB from infoteam[63]) as a basis to develop their own software framework called iLAB-Bio. The iLAB software framework is able to communicate with SiLA and OPC interface protocols generating their own generic driver for each device. The communication with both emerging standard protocols on the market (SiLA and OPC) is making it a good middleware framework for a digitalized laboratory. Using this framework, they were able to integrate a liquid handling platform using SiLA as the device communication protocol. On top of that, they used SiLA-based communication to integrate a bioreactor system with forty-eight 10 mL reaction vessels for parallelization and miniaturization. They also integrated the design of experiment (DoE) software MODDE into their digital infrastructure. For this, they used a combination of SiLA-based communication and manually matching process data to MODDE responses. While this method includes proprietary data formats, it still improves the data exchange with the software, decreasing the possibilities of data loss or damage.[9]
Seidel et al. and Poongothai et al. used various communication protocols (like SiLA2, OPC UA, gRPC, MQTT, or REST) to integrate laboratory devices into their comprehensive IT platform in an IoT fashion. Integrating devices using different interface options increases the maintenance cost and time, as well as the complexity, but this also allows the infrastructure to stay as flexible as possible. Automatically storing all available data in one place saves time, ensures data transparency, and reduces human error.[28]
Austerjost et al. also used the concept of IoT while using proprietary commands for laboratory device control.[34] They generated a digital device shadow for each device which was synchronized with the real laboratory device using the cloud-based IoT broker AWS IoT. The modular device integration was realized using the programming tool Node-RED[64], deployed on a Windows PC with both devices serially connected to the PC. The serial communication was possible with proprietary port commands that were provided by the vendor. The modular setup allows more device integration in the future, but using proprietary elements in the digitalization process will not lead to a universal solution for digitalized laboratories.[26][34][43]
In 2021, Porr et al. developed a digital infrastructure where device integration was achieved using SiLA-based communication.[35] The underlying concept of this solution was already published by Porr et al. in 2019.[33] Ethernet, USB, and RS232 is used to connect the (legacy) devices[45] with a wire making digital communication more stable than wireless communication due to less signal interfering possibilities. The infrastructure is based on a server-client-logic. It includes a process control system as their central process intelligence. Here, workflows, the sequence of steps required to complete the bioprocess, are planned and controlled. The process control system is connected to the aforementioned DeviceLayer, which communicates with the laboratory devices. After creating a workflow with the process control system using an intuitive GUI, the user can be guided through the laboratory work with the help of smartglasses as a NUI. The smartglasses show, for example, the next step of a complex assay that was incorporated into the workflow. Here it is important that this additional information is helping rather than distracting the user.[33] The use of smartglasses is just one example of using assistive devices in the laboratory, which will be further examined in the next subsection.
Laboratory assistants
Next to digitalizing laboratory devices and thus enabling digital communication, there are also other means for digital assistance in the laboratory. They can be distinguished into digital and physical assistants (Figure 4), but for both groups, their main goal is to help and support the person working in the laboratory. When working with laboratory assistants, it is unavoidable to address issues like privacy and confidentiality beforehand. Encrypted connections are an important factor for secure file transport. Other security methods like authentication, usage of (voice-)fingerprints, logging of system activities, message signing, or version control are becoming equally relevant when working with digital and physical assistants in digitalized laboratories.[28][34][36][43][50][52][57][65]
|
Digital assistants
Digital assistants include everything that might help the person working in the laboratory in their digital work, such as workflow scheduling, data gathering, or documenting experiments. The first digital assistant gaining importance in academic laboratories is the ELN. Requirements for the ELN are becoming more and more sophisticated and complex. It is not just about digital documentation of experiments but also about automated data gathering, connectivity to laboratory devices, sharing progress in team projects, as well as improving user-friendliness, data security, research quality.[25][66] With the ongoing digitalization of academic laboratories, most data are already digitally available and can be linked, copied, or integrated into the ELN. With the increasing availability of ELNs with different sizes and scopes, as well as increasing need for ELNs becoming evident, it is only a matter of time until all laboratories (including academia) will use an ELN instead of an analog laboratory notebook.[67][68] When switching an entire laboratory or institute to ELN, IT support needs to be considered: data might get stored on servers that need to be maintained; mobile devices, computers, and WIFI needs to be available; and upgrade and backup-support might be needed as well. After switching to an ELN, most users are preferring it over the paper-based version.[67] When going through the transformation towards a digitalized laboratory, it is important to take concerns raised by future users serious to increase acceptance of new tools and this new way of working in the laboratory.
The next level of digital assistance in bioprocess development laboratories is using a LIMS. The LIMS is where all data are collected, processed, managed, represented, and stored. The ELN is, in most cases, integrated into this management system. Using LIMS will give the laboratory user the advantage to digitally combine various parts of the laboratory such as data gathering, inventory management, workflow scheduling, sample tracking, data mining, device controlling, report creation, information overview, and metadata analysis.[28][53][67] Just as ELNs, there are various LIMS that differ in their scope and capabilities.[65] LIMS is also the basis of starting to enable model-based control of bioprocesses and automating workflows.[7] Another mean of digital assistance in the lab is the use of smart assistants like Amazon's Alexa. Laboratory workers can ask simple questions about melting points, molecular weight, and isoelectric points, or get answers to simple calculations during a workflow without having to physically interrupt their experiment.[69][70]
Physical assistants
Liquid handling stations, autosamplers, and real-time imaging devices are physically helping researchers with laboratory work. In recent years, however, stationary and mobile robots, as well as cobots, are starting to appear sporadically in academic laboratories supporting workers with various physical tasks.[71] Stationary robots are usually fixed to a place in the laboratory, unable to change their location by themselves, thus being mainly used to execute clear and repetitive workflows. Mounting them on rails gives them added spatial flexibility, increasing their potential application possibilities.[8] Mobile robots are taking on the task of transporting objects or samples inside the laboratory using their sensors, actuators, and information processing for navigation.[8][62] These mobile robots might even be drones in the future that use the air space in laboratories as transportation routes.[8] They are independent from human cooperation, mostly needing input or commands to start but then work on their own. The development of sensors and also the setup of the whole laboratory or building needs to be considered when using mobile robots. When they transport an object to a different floor, there needs to be some kind of elevator, automatic doors, and no unplanned obstacles in the way.[31]
Cobots on the other hand are collaborative robots that can be used in cooperation with humans and, due to their light weight design and improvement of safety features, they can also be used close to humans.[8][72] Robots in general are not common in academic laboratories yet, but researchers are starting to see the advantages that robots (stationary and mobile) and cobots offer. They rule out human volatility errors, do not mislabel samples, always transfer the right liquid or the right sample to the intended place, and will always log the right parameters.[12] Long development times, high maintenance needs, high monetary investments, and even reconstruction of whole laboratory areas are hindering the progress of increasing the amount of physical help in form of robots in academic laboratories. Additionally, in order for robots to be implemented into the academic laboratory, a digital infrastructure with a suited LIMS needs to be implemented first.[62] However, using robotics in the laboratory is helping to automate workflows or even whole processes, which will be further discussed in the following subsection.
Automated laboratories
A high level of automation is possible for a few laboratories in the field of academic bioprocess development. However, automation and high-throughput is not possible for most experimental settings in research laboratories due to the need of high flexibility and high variety of process workflows.[8] The complexity of academic laboratories is also justified in the high diversity of laboratory staff composition. It is not rare that personnel from more than one research project are sharing laboratories and or devices resulting in a different use of those. More challenges on the way to automating workflows are organizing the setup and ordering system[73], integrating standalone devices that are already automated by themselves[9], matching software requirements like legacy technology, proprietary interfaces, and new standard communication protocols (SiLA2, OPC UA).[9]
To collect as much data as possible for optimization of a bioprocess, Schmid et al. automated a setup for a DoE for cultivation experiments.[9] Using iLAB as the software framework, they integrated a liquid handling station and the DoE software MODDE into their digital infrastructure. The process parameters of cultivation experiments performed by two different bioreactor systems (96-well plate and 10 mL scale) were gathered and analyzed automatically in order to optimize the bioprocess. The integration of the liquid handling station and the standalone software MODDE, which was used to plan the experiments, was realized using SiLA2 and an iLAB plugin. This highly specific automated workflow made it possible to compare data and results from variously scaled experiments due to the focus on data integrity throughout the entire automated process.[9]
In the laboratory group of Buzz Baum at the University College London, Jennifer Rohn set up an automated microscopy for high-throughput screening of genes. Using an automated liquid handling system, short interfering RNA (siRNA), and automated microscopy for imaging, they are now able to screen genome-wide for genes that play a role in different morphological properties of human cells. After they overcame struggles to set up the workflow in the beginning, considering basic needs like freezer space or inventory control, they also found it challenging to find a good algorithm for analyzing the cell morphology. Using the automated setup saved their group a lot of time and made it possible to exploit a lot more of the available technology.[73]
Seidel et al. developed a comprehensive IT infrastructure for data handling and device integration.[28] Thus, they were able to automate a bioprocess workflow from online selection of enzymes through ordering gene sequences, cloning, transformation, cultivation, and preproduction to the final scale up to production scale. They focused on using a software that is comprehensive and easy to understand. Human error and workload of laboratory personnel as well as bioprocess developing times are reduced. Even though data storage and sharing is improved using their IT infrastructure, maintenance costs for databases, devices, and interfaces have increased. The whole digital infrastructure is only user-friendly as long as it helps the laboratory personnel in their tasks rather than making it more complicated.[28]
The potential of fully automated workflows or even fully automated laboratories in academic bioprocess development laboratories is yet to be discovered and will also not be possible everywhere due to the need of high flexibility and complexity. Dividing whole processes into sub-processes, developing a digital twin, using a modular setup, and staying flexible throughout the planning will ultimately help to automate an entire workflow.[7][8][28][74][75]
Artificial intelligence and machine learning
Artificial intelligence (AI) is a general term for the idea of code or algorithms to understand data in a way a human would, or carry out complex tasks based on data. Using AI tools like machine learning (ML) to analyze collected data can help to generate predictions or knowledge from big datasets efficiently. Besides analyzing simulated experiment data, AI and ML can also be used to solve challenges in everyday laboratory work. Especially when it comes to classification of data sets, protein structure predictions, or image recognition, these increasingly common techniques can prove useful.[76][77]
In 2021, Jumper et al. published their AI-based program AlphaFold that uses ML to predict the three-dimensional structure of proteins, based on their amino acid sequence, with a high accuracy.[78] AI-ML applications can help to make process control decisions that will impact the quality of the target product, as Nikita et al. showed in their work on continuous manufacturing of monoclonal antibodies.[77] While ML methods helped Volpato et al. to predict the enzymatic class of a protein based solely on the full amino acid sequence, other works showed the power of ML for classification problems as well. Austerjost et al. developed a mobile application for an image-based counting algorithm for Escherichia coli colonies on agar plates. Their idea was to use ML in order to have a more efficient sample throughput with high accuracy.[57] Biermann et al. used an algorithm-based image processing for the at-line determination of endospores from microscopic pictures.[79] Here, too, the aim was to achieve a faster result using ML. Thus, the presented applications are reducing the time to determine a certain number (E. coli colonies and endospores, respectively), making everyday laboratory work faster and more efficient.
Further and deeper information of ML in the field of bioprocess engineering can be read in the work of Duong-Trung et al.[80] As shown in these examples, AI or ML can make laboratory work faster and more efficient. To achieve these results, large amounts of data needed to be generated or harvested from online databases first. This data then needs to be explored, preprocessed, and or annotated manually. Here, education on how to handle big sets of data is beneficial for scientific staff. After data preparation models, neural networks or algorithms need to be developed, trained, and optimized before they can provide the support the laboratory worker is anticipating.[76] Therefore, to unlock the whole potential of AI or ML in the laboratory, digitalization for automated secure data capture is needed as the first step.
Simulation of experiments using process models
Digital twins, as previously discussed, have, among other things, potential to gather great amounts of data due to their real-time data monitoring. With advancing digitalization in bioprocess laboratories, the amount of data from experiments is therefore also increasing. Based upon this obtained data, process models can be designed that represent the biotechnological process as a whole to run simulations, with the aim to extract new valuable data.[11][81][82] Modeling experiments and simulating their respective outcomes using parameter adjustment is unlocking new potential to get information without having to do all the actual work inside the laboratory.[76] After a process model has been developed, it is important to verify its accuracy either through experimental validation experiments or through expert knowledge.[7]
Zobel-Roos et al. used the data of only a small amount of experiments for a process model to evaluate the outcome of different continuous chromatography setups. These small experiments were done to estimate the parameters for the process model. The results determined by the model were then experimentally verified in the laboratory. Simulation and tracer experiments showed a deviation as small as 0.8%.[83] Ladd Effio et al. showed successful process modeling approaches for the downstream part of a bioprocess optimizing anion-exchange chromatography. After experimentally determining adsorption isotherms and mass transfer parameters they were subsequently able to use their simulation-based model to identify optimal elution conditions.[84] Boi et al. were able to develop a model of affinity membrane chromatography to help with the prediction of membrane performance in bioprocess design.[85]
Cell type, physiology, metabolism, and more factors are substantially increasing the complexity of bioprocesses when whole cells are involved. While representing a reliable mathematical model of a whole cell is still seemingly impossible, model-based methods are able to make predictions. These predictions cannot be fully trusted and have to be verified with experiments in the laboratory. However, designing bioprocesses by analyzing data with ML methods and simulations can help to optimize experimental setups that can subsequently be transferred to the laboratory.[2][7]
Virtual laboratories
Besides digitalizing the physical laboratory, there is another approach that is particularly practical for educational purposes: virtual laboratories. The concept of these laboratories is that results of experiments are being photographed or otherwise documented and stored with their respective settings. These pictures and information are then uploaded to the virtual laboratory (online database). As depicted in Figure 5, users can then access this virtual laboratory through, for example, a graphical web-service, alternating the settings of the virtual experimental setup and receiving the associated picture or outcome.[86] This tool proves to be useful for a deeper understanding of complex device equipment. A study found that students who completed an online device training in a virtual laboratory before conducting the experiment in the physical lab needed less guidance and were able to spend more time for the actual experiment rather than getting familiar with the complex equipment. Having learned the basic equipment knowledge online, these students were better prepared and more confident than those who did not use the virtual laboratory beforehand.[87][88] When students use virtual instead of traditional laboratories for educational purposes, there is no difference in the effectiveness of content knowledge.[89] The easy and automated generation of virtual laboratories is well described in the work of De La Torre et al.[90] Even if this digitalization strategy has no impact for academic laboratories to generate new research data, this can well be used for training or educational purposes.[90]
|
The use of virtual reality (VR) is another way to interpret the term virtual laboratories. For better understanding, the terms "VR" and "augmented reality" (AR) are shortly explained by Nakhle et al.[91]:
- Virtual reality: “Immerses users into interactive and explorative virtual environments based on 3D representation and simulation of the real world and objects within it.” Requirements: VR headset, PC.[91]
- Augmented reality: “Superimposes valuable visual content and information over a live view of the real world to provide users an enriched version of reality, allowing them to interact with digital objects in real-time.” Requirements: Smartphone & tablet, AR glasses.[91]
Harfouche et al. showed that when studying biotechnology, bioethics can be taught via VR, making a virtual classroom experience possible for students.[92] Another example for education via VR is the Arizona State University, which is offering a biology degree solely based on VR laboratory work.[88][93] Advantages of these VR classrooms or laboratories include the student's local independence, no exposure to toxic chemicals, no spatial limitations, the possibility to slow down very fast reactions for visualization, and no safety concerns when learning about, for example, explosives or toxins. Thus, the learning experience using VR will be enhanced in a cost and time effective manner. Additionally, students will interactively be taught with reduced human bias regarding gender, age, ethnicity, and race.[88][91][92]
VR can be a helpful tool for scientists to visualize 3D images to grasp cell's morphological features with confocal microscopy. ConfocalVR is a VR application that can be installed on consumer-grade VR systems to visualize microscopic image stacks into 3D images in the virtual world. The user can then interact with the digital object and examine its properties in a way that would not be possible with 2D imaging on a computer screen.[94]
Device training and or maintenance can be improved by using AR. Device experts do not have to travel to laboratories but can guide the scientists through maintenance procedures with instructions while being able to see what the scientists who wears AR glasses sees. Instructions and information can be displayed in the user's view for guidance. This way requesting assistance from local independent experts, teachers, or colleagues is not more than a call away linking more knowledge together.[95][96][97]
Conclusions
Digitalizing academic bioprocess development laboratories can lead to a new transformed era of research. When supported by digital assistants, using connected devices, more intuitive user interfaces, and data gathering according to FAIR data principles, academic research will become less time consuming and more effective and accurate, as well as less prone to (human) error. This will also lead to a better reproducibility of experiments due to improved data integrity and availability. Integrating devices into digital infrastructures is becoming easier because of the increased use of standard communication protocols like SiLA2 and LADS. New students can be prepared for these new kinds of digital laboratories with proper education, including learning basic IT skills, like scripting knowledge or the ability to handle big data sets.
Digital or physical assistants can be a good way to help personnel working in the laboratory, but their implementation and maintenance is complex. Collaborating with IT experts will help to overcome this issue, as well as the task of seamless integration and communication of laboratory devices. When working in automated laboratories, IT experts working closely with laboratory workers are crucial for creating and executing automated research workflows. Communication between scientific researchers and IT experts will become essential because even the most sophisticated (from the IT perspective) digitalization concept will be useless when it complicates the laboratory work. This communication is only possible if both sides have a basic vocabulary and understanding of the other topic. Simulating experiments using process models have potential to decrease laboratory work while shifting the work to IT experts, who also have to understand the basic science behind the experiment. Besides IT experts with experimental knowledge, laboratory staff with IT knowledge will advance the highly interdisciplinary process of digitalization as well.
Virtual laboratories are good for training or educational purposes but will not produce any new scientifically relevant data. Integrating VR or AR in laboratory work or laboratory education programs is a cost-effective way of protecting scientific staff and or students. Besides, the possibility of new points of view on experiments, cells, or other small particles or fast processes can add new value to research.
Finally, there is not a single digitalization strategy that fits all. The needs and requirements, as well as the level of flexibility and complexity, is very broad in the field of academic bioprocess development. Thus, quality collaboration and communication between IT experts and laboratory staff, consideration of standard device communication protocols, following FAIR data principles, and making sure that the solution supports the laboratory worker rather than disturb them will lead to a maintainable digital infrastructure fitting researchers' individual needs. Additions to the education of new scientists and interdisciplinary networking is also a key aspect to drive this transformation towards digitalized laboratories in the academic field of bioprocess development.
Abbreviations, acronyms, and initialisms
- AI: artificial intelligence
- AR: augmented reality
- CLI: command line interface
- DoE: design of experiments
- ELN: electronic laboratory notebook
- FAIR: findable, accessible, interoperable, reusable
- GUI: graphical user interface
- IoT: internet of things
- LADS: Laboratory and Analytical Device Standard
- LIMS: laboratory information management system
- ML: machine learning
- NUI: natural user interface
- OPC UA: open platform communications unified architecture
- SiLA2: Standardization in Lab Automation 2
- VR: virtual reality
- VUI: voice user interface
Acknowledgements
The authors thank the German Ministry of Education and Research (BMBF) for funding within the National Joint Research Project Digitalization in Industrial Biotechnology (DigInBio). Grant number: FKZ 031B0463B. The authors would like to thank the Open Access fund of Leibniz University Hannover for the funding of the publication of this article. T.H. would like to thank Ferdinand Lange for his help with technical questions.
Open access funding enabled and organized by Projekt DEAL.
Data availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Conflict of interest
The authors declare no conflicts of interest.
References
- ↑ 1.0 1.1 Hollricher, K. (2022). "Digitales Lab - Viel luft nach oben". Carl 1: 42–5. https://sila-standard.com/wp-content/uploads/2022/11/Digitales-Lab-viel-Luft-nach-oben.pdf.
- ↑ 2.0 2.1 2.2 Neubauer, Peter; Glauche, Florian; Cruz‐Bournazou, M. Nicolas (1 November 2017). "Editorial: Bioprocess Development in the era of digitalization" (in en). Engineering in Life Sciences 17 (11): 1140–1141. doi:10.1002/elsc.201770113. ISSN 1618-0240. PMC PMC6999283. PMID 32624741. https://onlinelibrary.wiley.com/doi/10.1002/elsc.201770113.
- ↑ Jochums, Max; Reinders, Lars M. H.; Tuerk, Jochen; Teutenberg, Thorsten (2022), Beutel, Sascha; Lenk, Felix, eds., "Flexible Digitization of Highly Individualized Workflows Demonstrated Through the Quality Control of Patient-Specific Cytostatic Application Bags: Digitization from the Perspective of Small and Medium-Sized Laboratories" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 115–129, doi:10.1007/10_2021_190, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2021_190. Retrieved 2024-04-23
- ↑ Lütjohann, Dominic S.; Jung, Nicole; Bräse, Stefan (1 May 2015). "Open source life science automation: Design of experiments and data acquisition via “dial-a-device”" (in en). Chemometrics and Intelligent Laboratory Systems 144: 100–107. doi:10.1016/j.chemolab.2015.04.002. https://linkinghub.elsevier.com/retrieve/pii/S0169743915000842.
- ↑ 5.0 5.1 Kockmann, Norbert (2019). "Digital methods and tools for chemical equipment and plants" (in en). Reaction Chemistry & Engineering 4 (9): 1522–1529. doi:10.1039/C9RE00017H. ISSN 2058-9883. https://xlink.rsc.org/?DOI=C9RE00017H.
- ↑ Krüger, Anna; Schäfers, Christian; Busch, Philip; Antranikian, Garabed (1 November 2020). "Digitalization in microbiology – Paving the path to sustainable circular bioeconomy" (in en). New Biotechnology 59: 88–96. doi:10.1016/j.nbt.2020.06.004. https://linkinghub.elsevier.com/retrieve/pii/S1871678420301497.
- ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 7.10 Richter, Jannik; Lange, Ferdinand; Scheper, Thomas; Solle, Dörte; Beutel, Sascha (1 April 2023). "Digitale Zwillinge in der Bioprozesstechnik – Chancen und Möglichkeiten" (in en). Chemie Ingenieur Technik 95 (4): 498–510. doi:10.1002/cite.202200166. ISSN 0009-286X. https://onlinelibrary.wiley.com/doi/10.1002/cite.202200166.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 8.8 Thurow, Kerstin (2021), Beutel, Sascha; Lenk, Felix, eds., "Automation for Life Science Laboratories" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 3–22, doi:10.1007/10_2021_170, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2021_170. Retrieved 2024-04-23
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 9.8 9.9 Schmid, Ingrid; Aschoff, Joachim (1 November 2017). "A scalable software framework for data integration in bioprocess development" (in en). Engineering in Life Sciences 17 (11): 1159–1165. doi:10.1002/elsc.201600008. ISSN 1618-0240. PMC PMC6999508. PMID 32624743. https://onlinelibrary.wiley.com/doi/10.1002/elsc.201600008.
- ↑ Neubauer, Peter; Cruz, Nicolas; Glauche, Florian; Junne, Stefan; Knepper, Andreas; Raven, Michael (1 May 2013). "Consistent development of bioprocesses from microliter cultures to the industrial scale" (in en). Engineering in Life Sciences 13 (3): 224–238. doi:10.1002/elsc.201200021. ISSN 1618-0240. https://onlinelibrary.wiley.com/doi/10.1002/elsc.201200021.
- ↑ 11.0 11.1 Narayanan, Harini; Luna, Martin F.; von Stosch, Moritz; Cruz Bournazou, Mariano Nicolas; Polotti, Gianmarco; Morbidelli, Massimo; Butté, Alessandro; Sokolov, Michael (1 January 2020). "Bioprocessing in the Digital Age: The Role of Process Models" (in en). Biotechnology Journal 15 (1): 1900172. doi:10.1002/biot.201900172. ISSN 1860-6768. https://onlinelibrary.wiley.com/doi/10.1002/biot.201900172.
- ↑ 12.0 12.1 12.2 Saha, Himadri Nath; Auddy, Supratim; Pal, Subrata; Kumar, Shubham; Jasu, Subhadeep; Singh, Rocky; Singh, Rakhee; Banerjee, Swarnadeep et al. (1 August 2017). "Internet of Things (IoT) on bio-technology". 2017 8th Annual Industrial Automation and Electromechanical Engineering Conference (IEMECON) (Bangkok, Thailand: IEEE): 364–369. doi:10.1109/IEMECON.2017.8079624. ISBN 978-1-5386-2215-5. http://ieeexplore.ieee.org/document/8079624/.
- ↑ Osthege, Michael; Tenhaef, Niklas; Zyla, Rebecca; Müller, Carolin; Hemmerich, Johannes; Wiechert, Wolfgang; Noack, Stephan; Oldiges, Marco (1 March 2022). "bletl ‐ A Python package for integrating BioLector microcultivation devices in the Design‐Build‐Test‐Learn cycle" (in en). Engineering in Life Sciences 22 (3-4): 242–259. doi:10.1002/elsc.202100108. ISSN 1618-0240. PMC PMC8961055. PMID 35382539. https://onlinelibrary.wiley.com/doi/10.1002/elsc.202100108.
- ↑ Solle, Dörte; Hitzmann, Bernd; Herwig, Christoph; Pereira Remelhe, Manuel; Ulonska, Sophia; Wuerth, Lynn; Prata, Adrian; Steckenreiter, Thomas (1 May 2017). "Between the Poles of Data‐Driven and Mechanistic Modeling for Process Operation" (in en). Chemie Ingenieur Technik 89 (5): 542–561. doi:10.1002/cite.201600175. ISSN 0009-286X. https://onlinelibrary.wiley.com/doi/10.1002/cite.201600175.
- ↑ Wong, Sybil; Pan, Mingke; Shaw, Allen; Gershater, Markus (1 July 2022). "What digitalization in biology R&D means for biotech companies and life scientists" (in en). Nature Biotechnology 40 (7): 1151–1153. doi:10.1038/s41587-022-01309-y. ISSN 1087-0156. https://www.nature.com/articles/s41587-022-01309-y.
- ↑ Weiss, Charles J. (9 May 2017). "Scientific Computing for Chemists: An Undergraduate Course in Simulations, Data Processing, and Visualization" (in en). Journal of Chemical Education 94 (5): 592–597. doi:10.1021/acs.jchemed.7b00078. ISSN 0021-9584. https://pubs.acs.org/doi/10.1021/acs.jchemed.7b00078.
- ↑ 17.0 17.1 17.2 17.3 17.4 17.5 Kraus, V.P. (11 October 2021). "Von OPC UA zu LADS: Ein Kommunikationsstandard entsteht". Labor Praxis. Vogel Communications Group. https://www.laborpraxis.vogel.de/von-opc-ua-zu-lads-ein-kommunikationsstandard-entsteht-a-13c637c18db4e91c678b4734c0c6b941/.
- ↑ 18.0 18.1 European Commission. Directorate General for Research and Innovation. (2018). Turning FAIR into reality: final report and action plan from the European Commission expert group on FAIR data.. LU: Publications Office. doi:10.2777/1524. https://data.europa.eu/doi/10.2777/1524.
- ↑ Landi, Annalisa; Thompson, Mark; Giannuzzi, Viviana; Bonifazi, Fedele; Labastida, Ignasi; da Silva Santos, Luiz Olavo Bonino; Roos, Marco (1 January 2020). "The “A” of FAIR – As Open as Possible, as Closed as Necessary" (in en). Data Intelligence 2 (1-2): 47–55. doi:10.1162/dint_a_00027. ISSN 2641-435X. https://direct.mit.edu/dint/article/2/1-2/47-55/9998.
- ↑ Mons, Barend; Neylon, Cameron; Velterop, Jan; Dumontier, Michel; da Silva Santos, Luiz Olavo Bonino; Wilkinson, Mark D. (7 March 2017). "Cloudy, increasingly FAIR; revisiting the FAIR Data guiding principles for the European Open Science Cloud". Information Services & Use 37 (1): 49–56. doi:10.3233/ISU-170824. https://www.medra.org/servlet/aliasResolver?alias=iospress&doi=10.3233/ISU-170824.
- ↑ 21.0 21.1 Solle, Dörte (1 July 2020). "Be FAIR to your data" (in en). Analytical and Bioanalytical Chemistry 412 (17): 3961–3965. doi:10.1007/s00216-020-02526-7. ISSN 1618-2642. PMC PMC7320032. PMID 32300841. https://link.springer.com/10.1007/s00216-020-02526-7.
- ↑ Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, IJsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship" (in en). Scientific Data 3 (1): 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. PMC PMC4792175. PMID 26978244. https://www.nature.com/articles/sdata201618.
- ↑ Atici, Levent; Kansa, Sarah Whitcher; Lev-Tov, Justin; Kansa, Eric C. (1 December 2013). "Other People’s Data: A Demonstration of the Imperative of Publishing Primary Data" (in en). Journal of Archaeological Method and Theory 20 (4): 663–681. doi:10.1007/s10816-012-9132-9. ISSN 1072-5369. http://link.springer.com/10.1007/s10816-012-9132-9.
- ↑ Neumann, Janna (2022), Beutel, Sascha; Lenk, Felix, eds., "FAIR Data Infrastructure" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 195–207, doi:10.1007/10_2021_193, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2021_193. Retrieved 2024-04-23
- ↑ 25.0 25.1 Dunie, Matt (7 November 2017). Lawlor, Bonnie. ed. "The importance of research data management: The value of electronic laboratory notebooks in the management of data integrity and data availability". Information Services & Use 37 (3): 355–359. doi:10.3233/ISU-170843. https://www.medra.org/servlet/aliasResolver?alias=iospress&doi=10.3233/ISU-170843.
- ↑ 26.0 26.1 26.2 26.3 26.4 Gauglitz, Günter (1 August 2018). "Lab 4.0: SiLA or OPC UA" (in en). Analytical and Bioanalytical Chemistry 410 (21): 5093–5094. doi:10.1007/s00216-018-1192-6. ISSN 1618-2642. http://link.springer.com/10.1007/s00216-018-1192-6.
- ↑ Hinkel, Georg; Kunert, Jörg; Meredith, Jason (1 October 2023). "The Tecan SiLA2 SDK: A royalty-free, open-source framework to develop SiLA2 servers and clients" (in en). SLAS Technology 28 (5): 334–344. doi:10.1016/j.slast.2023.07.001. https://linkinghub.elsevier.com/retrieve/pii/S2472630323000444.
- ↑ 28.0 28.1 28.2 28.3 28.4 28.5 28.6 28.7 Seidel, Simon; Cruz-Bournazou, Mariano Nicolas; Groß, Sebastian; Schollmeyer, Julia Katharina; Kurreck, Anke; Krauss, Stefan; Neubauer, Peter (2022), Beutel, Sascha; Lenk, Felix, eds., "A Comprehensive IT Infrastructure for an Enzymatic Product Development in a Digitalized Biotechnological Laboratory" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 61–82, doi:10.1007/10_2022_207, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2022_207. Retrieved 2024-04-23
- ↑ Bai, Jiaru; Cao, Liwei; Mosbach, Sebastian; Akroyd, Jethro; Lapkin, Alexei A.; Kraft, Markus (28 February 2022). "From Platform to Knowledge Graph: Evolution of Laboratory Automation" (in en). JACS Au 2 (2): 292–309. doi:10.1021/jacsau.1c00438. ISSN 2691-3704. PMC PMC8889618. PMID 35252980. https://pubs.acs.org/doi/10.1021/jacsau.1c00438.
- ↑ 30.0 30.1 30.2 30.3 Freundel, Matthias (2022), Beutel, Sascha; Lenk, Felix, eds., "Comparison of Laboratory Standards" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 133–145, doi:10.1007/10_2022_205, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2022_205. Retrieved 2024-04-23
- ↑ 31.0 31.1 Wolf, Ádám; Wolton, David; Trapl, Josef; Janda, Julien; Romeder-Finger, Stefan; Gatternig, Thomas; Farcet, Jean-Baptiste; Galambos, Péter et al. (1 February 2022). "Towards robotic laboratory automation Plug & Play: The “LAPP” framework" (in en). SLAS Technology 27 (1): 18–25. doi:10.1016/j.slast.2021.11.003. https://linkinghub.elsevier.com/retrieve/pii/S2472630321000224.
- ↑ Schmieder, Florian; Polk, Christoph; Gottlöber, Felix; Schöps, Patrick; Sonntag, Frank; Deuse, Ronny; Jede, Aline; Petzold, Thomas (1 September 2019). "Universal LIMS based platform for the automated processing of cell-based assays" (in en). Current Directions in Biomedical Engineering 5 (1): 437–439. doi:10.1515/cdbme-2019-0110. ISSN 2364-5504. https://www.degruyter.com/document/doi/10.1515/cdbme-2019-0110/html.
- ↑ 33.0 33.1 33.2 Porr, Marc; Marquard, Daniel; Stanislawski, Nils; Austerjost, Jonas; Russo, Mario; Bungers, Simon; Klimmt, Christoph; Scheper, Thomas et al. (1 March 2019). "smartLAB – Interaktives Arbeiten in digitalisierter Laborumgebung" (in en). Chemie Ingenieur Technik 91 (3): 285–293. doi:10.1002/cite.201800090. ISSN 0009-286X. https://onlinelibrary.wiley.com/doi/10.1002/cite.201800090.
- ↑ 34.0 34.1 34.2 34.3 34.4 Austerjost, Jonas; Porr, Marc; Riedel, Noah; Geier, Dominik; Becker, Thomas; Scheper, Thomas; Marquard, Daniel; Lindner, Patrick et al. (1 October 2018). "Introducing a Virtual Assistant to the Lab: A Voice User Interface for the Intuitive Control of Laboratory Instruments" (in en). SLAS Technology 23 (5): 476–482. doi:10.1177/2472630318788040. https://linkinghub.elsevier.com/retrieve/pii/S2472630322012651.
- ↑ 35.0 35.1 35.2 35.3 35.4 Porr, Marc; Lange, Ferdinand; Marquard, Daniel; Niemeyer, Laura; Lindner, Patrick; Scheper, Thomas; Beutel, Sascha (1 March 2021). "Implementing a digital infrastructure for the lab using a central laboratory server and the SiLA2 communication standard" (in en). Engineering in Life Sciences 21 (3-4): 208–219. doi:10.1002/elsc.202000053. ISSN 1618-0240. PMC PMC7923558. PMID 33716619. https://onlinelibrary.wiley.com/doi/10.1002/elsc.202000053.
- ↑ 36.0 36.1 36.2 36.3 Juchli, Daniel (2022), Beutel, Sascha; Lenk, Felix, eds., "SiLA 2: The Next Generation Lab Automation Standard" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 147–174, doi:10.1007/10_2022_204, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2022_204. Retrieved 2024-04-23
- ↑ Ktori, S. (May 2021). "SiLA for Instrument Vendors". SiLA Analyzed 1: 1–4. https://sila-standard.com/wp-content/uploads/2021/06/SiLA-for-instrument-vendors-WhitePaper.pdf.
- ↑ 38.0 38.1 38.2 "SiLA Frequently Asked Questions". SiLA Consortium. https://sila-standard.com/faq/. Retrieved 02 September 2023.
- ↑ "Unified Architecture". OPC Foundation. https://opcfoundation.org/about/opc-technologies/opc-ua/.
- ↑ 40.0 40.1 40.2 Bär, Henning; Hochstrasser, Remo; Papenfuß, Bernd (1 April 2012). "SiLA: Basic Standards for Rapid Integration in Laboratory Automation" (in en). SLAS Technology 17 (2): 86–95. doi:10.1177/2211068211424550. https://linkinghub.elsevier.com/retrieve/pii/S2472630322016673.
- ↑ Rachuri, Sudarsan; Subrahmanian, Eswaran; Bouras, Abdelaziz; Fenves, Steven J.; Foufou, Sebti; Sriram, Ram D. (1 July 2008). "Information sharing and exchange in the context of product lifecycle management: Role of standards" (in en). Computer-Aided Design 40 (7): 789–800. doi:10.1016/j.cad.2007.06.012. https://linkinghub.elsevier.com/retrieve/pii/S0010448507001613.
- ↑ 42.0 42.1 42.2 42.3 42.4 Bromig, Lukas; Leiter, David; Mardale, Alexandru-Virgil; von den Eichen, Nikolas; Bieringer, Emmeran; Weuster-Botz, Dirk (1 January 2022). "The SiLA 2 Manager for rapid device integration and workflow automation" (in en). SoftwareX 17: 100991. doi:10.1016/j.softx.2022.100991. https://linkinghub.elsevier.com/retrieve/pii/S2352711022000103.
- ↑ 43.0 43.1 43.2 43.3 Brendel, Alexander; Dorfmüller, Franziska; Liebscher, Albrecht; Kraus, Patrick; Kress, Kai; Oehme, Heiko; Arnold, Matthias; Koschitzki, Robert (2022), Beutel, Sascha; Lenk, Felix, eds., "Laboratory and Analytical Device Standard (LADS): A Communication Standard Based on OPC UA for Networked Laboratories" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 175–194, doi:10.1007/10_2022_209, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2022_209. Retrieved 2024-04-23
- ↑ 44.0 44.1 44.2 "Was ist OPC UA? Die wichtigsten Begriffe im Überblick". OPC Router. inray Industriesoftware GmbH. 2023. https://www.opc-router.de/was-ist-opc-ua/.
- ↑ 45.0 45.1 45.2 45.3 45.4 Porr, Marc; Schwarz, Sebastian; Lange, Ferdinand; Niemeyer, Laura; Hentrop, Thorleif; Marquard, Daniel; Lindner, Patrick; Scheper, Thomas et al. (1 October 2020). "Bringing IoT to the Lab: SiLA2 and Open-Source-Powered Gateway Module for Integrating Legacy Devices into the Digital Laboratory" (in en). HardwareX 8: e00118. doi:10.1016/j.ohx.2020.e00118. PMC PMC9041174. PMID 35498267. https://linkinghub.elsevier.com/retrieve/pii/S2468067220300274.
- ↑ "SiLA Downloads". SiLA Consortium. https://sila-standard.com/downloads/. Retrieved 14 April 2023.
- ↑ "SPECTARIS Standard for Laboratory Equipment Communication". OPC Connect. OPC Foundation. December 2020. https://opcconnect.opcfoundation.org/2020/12/spectaris-standard-for-laboratory-equipment-communication/.
- ↑ "LADS – Laboratory and Analytical Device Standard". OPC Foundation. https://opcfoundation.org/markets-collaboration/lads/.
- ↑ 49.0 49.1 Mahnke, Wolfgang; Leitner, Stefan-Helmut; Damm, Matthias (2009) (in en). OPC Unified Architecture. Berlin, Heidelberg: Springer Berlin Heidelberg. doi:10.1007/978-3-540-68899-0. ISBN 978-3-540-68898-3. http://link.springer.com/10.1007/978-3-540-68899-0.
- ↑ 50.0 50.1 Poongothai, M.; Subramanian, P. Muthu; Rajeswari, A. (1 April 2018). "Design and implementation of IoT based smart laboratory". 2018 5th International Conference on Industrial Engineering and Applications (ICIEA) (Singapore: IEEE): 169–173. doi:10.1109/IEA.2018.8387090. ISBN 978-1-5386-5747-8. https://ieeexplore.ieee.org/document/8387090/.
- ↑ Patel, K.K; Patel, S.M. (2016). "Internet of Things-IOT: Definition, Characteristics, Architecture, Enabling Technologies, Application & Future Challenges". International Journal of Engineering Science and Computing 6 (5): 6122–31. doi:10.4010/2016.1482. https://fardapaper.ir/mohavaha/uploads/2019/09/Fardapaper-Internet-of-Things-IOT-Definition-Characteristics-Architecture-Enabling-Technologies-Application-Future-Challenges.pdf.
- ↑ 52.0 52.1 Garg, Hittu; Dave, Mayank (1 April 2019). "Securing IoT Devices and SecurelyConnecting the Dots Using REST API and Middleware". 2019 4th International Conference on Internet of Things: Smart Innovation and Usages (IoT-SIU) (Ghaziabad, India: IEEE): 1–6. doi:10.1109/IoT-SIU.2019.8777334. ISBN 978-1-7281-1253-4. https://ieeexplore.ieee.org/document/8777334/.
- ↑ 53.0 53.1 53.2 Söldner, Robert; Rheinländer, Sophia; Meyer, Tim; Olszowy, Michael; Austerjost, Jonas (2022), Beutel, Sascha; Lenk, Felix, eds., "Human–Device Interaction in the Life Science Laboratory" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 83–113, doi:10.1007/10_2021_183, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2021_183. Retrieved 2024-04-23
- ↑ 54.0 54.1 54.2 Fuller, Aidan; Fan, Zhong; Day, Charles; Barlow, Chris (2020). "Digital Twin: Enabling Technologies, Challenges and Open Research". IEEE Access 8: 108952–108971. doi:10.1109/ACCESS.2020.2998358. ISSN 2169-3536. https://ieeexplore.ieee.org/document/9103025/.
- ↑ 55.0 55.1 55.2 Kritzinger, Werner; Karner, Matthias; Traar, Georg; Henjes, Jan; Sihn, Wilfried (2018). "Digital Twin in manufacturing: A categorical literature review and classification" (in en). IFAC-PapersOnLine 51 (11): 1016–1022. doi:10.1016/j.ifacol.2018.08.474. https://linkinghub.elsevier.com/retrieve/pii/S2405896318316021.
- ↑ 56.0 56.1 Wolf, Adam; Galambos, Peter; Szell, Karoly (1 July 2020). "Device Integration Concepts in Laboratory Automation". 2020 IEEE 24th International Conference on Intelligent Engineering Systems (INES) (Reykjavík, Iceland: IEEE): 171–178. doi:10.1109/INES49302.2020.9147171. ISBN 978-1-7281-1059-2. https://ieeexplore.ieee.org/document/9147171/.
- ↑ 57.0 57.1 57.2 Austerjost, Jonas; Marquard, Daniel; Raddatz, Lukas; Geier, Dominik; Becker, Thomas; Scheper, Thomas; Lindner, Patrick; Beutel, Sascha (1 August 2017). "A smart device application for the automated determination of E. coli colonies on agar plates" (in en). Engineering in Life Sciences 17 (8): 959–966. doi:10.1002/elsc.201700056. ISSN 1618-0240. PMC PMC6999497. PMID 32624845. https://onlinelibrary.wiley.com/doi/10.1002/elsc.201700056.
- ↑ Gan, Samuel Ken-En; Poon, Jun-Kai (1 December 2016). "The world of biomedical apps: their uses, limitations, and potential" (in en). Scientific Phone Apps and Mobile Devices 2 (1): 6. doi:10.1186/s41070-016-0009-2. ISSN 2364-4958. http://scientificphoneapps.springeropen.com/articles/10.1186/s41070-016-0009-2.
- ↑ Min, Hyun Jung; Mina, Hansel A.; Deering, Amanda J.; Bae, Euiwon (1 September 2021). "Development of a smartphone-based lateral-flow imaging system using machine-learning classifiers for detection of Salmonella spp." (in en). Journal of Microbiological Methods 188: 106288. doi:10.1016/j.mimet.2021.106288. https://linkinghub.elsevier.com/retrieve/pii/S0167701221001561.
- ↑ Tatman, Rachael; Kasten, Conner (20 August 2017). "Effects of Talker Dialect, Gender & Race on Accuracy of Bing Speech and YouTube Automatic Captions" (in en). Interspeech 2017 (ISCA): 934–938. doi:10.21437/Interspeech.2017-1746. https://www.isca-archive.org/interspeech_2017/tatman17_interspeech.html.
- ↑ Salazar Ch., Gustavo D.; Venegas, Carlos; Baca, Michael; Rodriguez, Ismael; Marrone, Luis (1 November 2018). "Open Middleware proposal for IoT focused on Industry 4.0". 2018 IEEE 2nd Colombian Conference on Robotics and Automation (CCRA) (Barranquilla: IEEE): 1–6. doi:10.1109/CCRA.2018.8588117. ISBN 978-1-5386-8464-1. https://ieeexplore.ieee.org/document/8588117/.
- ↑ 62.0 62.1 62.2 62.3 Biermann, Ferdinand; Mathews, Julius; Nießing, Bastian; König, Niels; Schmitt, Robert H. (29 May 2021). "Automating Laboratory Processes by Connecting Biotech and Robotic Devices—An Overview of the Current Challenges, Existing Solutions and Ongoing Developments" (in en). Processes 9 (6): 966. doi:10.3390/pr9060966. ISSN 2227-9717. https://www.mdpi.com/2227-9717/9/6/966.
- ↑ "Life Science". Inforteam Software. https://infoteam.de/unsere-maerkte/life-science.
- ↑ "Node-RED". OpenJS Foundation. https://nodered.org/.
- ↑ 65.0 65.1 Prasad, Poonam J.; Bodhe, G.L. (1 August 2012). "Trends in laboratory information management system" (in en). Chemometrics and Intelligent Laboratory Systems 118: 187–192. doi:10.1016/j.chemolab.2012.07.001. https://linkinghub.elsevier.com/retrieve/pii/S0169743912001438.
- ↑ Potthoff, Jan; Tremouilhac, Pierre; Hodapp, Patrick; Neumair, Bernhard; Bräse, Stefan; Jung, Nicole (1 March 2019). "Procedures for systematic capture and management of analytical data in academia" (in en). Analytica Chimica Acta: X 1: 100007. doi:10.1016/j.acax.2019.100007. PMC PMC7587027. PMID 33117974. https://linkinghub.elsevier.com/retrieve/pii/S2590134619300039.
- ↑ 67.0 67.1 67.2 Dirnagl, Ulrich; Przesdzing, Ingo (4 January 2016). "A pocket guide to electronic laboratory notebooks in the academic life sciences" (in en). F1000Research 5: 2. doi:10.12688/f1000research.7628.1. ISSN 2046-1402. PMC PMC4722687. PMID 26835004. https://f1000research.com/articles/5-2/v1.
- ↑ Voegele, Catherine; Bouchereau, Baptiste; Robinot, Nivonirina; McKay, James; Damiecki, Philippe; Alteyrac, Lucile (1 July 2013). "A universal open-source Electronic Laboratory Notebook" (in en). Bioinformatics 29 (13): 1710–1712. doi:10.1093/bioinformatics/btt253. ISSN 1367-4803. https://academic.oup.com/bioinformatics/article/29/13/1710/200295.
- ↑ Condliffe, J. (3 May 2017). "Scientists Are Turning Alexa into an Automated Lab Helper". MIT Technology Review. https://www.technologyreview.com/2017/05/03/68492/scientists-are-turning-alexa-into-an-automated-lab-helper/.
- ↑ Halford, B. (2 May 2017). "Meet your new lab assistant". Chemical & Engineering News 95 (19). https://cen.acs.org/articles/95/i19/Meet-your-new-lab-assistant.html.
- ↑ Liu, Hui; Stoll, Norbert; Junginger, Steffen; Thurow, Kerstin (1 July 2013). "Mobile Robot for Life Science Automation" (in en). International Journal of Advanced Robotic Systems 10 (7): 288. doi:10.5772/56670. ISSN 1729-8814. http://journals.sagepub.com/doi/10.5772/56670.
- ↑ Ghandour, M.; Liu, H.; Stoll, N. et al. (2016). "A hybrid collision avoidance system for indoor mobile robots based on human-robot interaction". Proceedings of the 17th International Conference on Mechatronics - Mechatronika (ME): 1–7. https://ieeexplore.ieee.org/document/7827795.
- ↑ 73.0 73.1 Blow, Nathan (1 January 2008). "Lab automation: tales along the road to automation" (in en). Nature Methods 5 (1): 109–112. doi:10.1038/nmeth0108-109. ISSN 1548-7091. https://www.nature.com/articles/nmeth0108-109.
- ↑ Schuh, Günther; Walendzik, Pia; Luckert, Melanie; Birkmeier, Martin; Weber, Anja; Blum, Matthias (28 November 2016). "Keine Industrie 4.0 ohne den Digitalen Schatten: Wie Unternehmen die notwendige Datenbasis schaffen" (in en). Zeitschrift für wirtschaftlichen Fabrikbetrieb 111 (11): 745–748. doi:10.3139/104.111613. ISSN 2511-0896. https://www.degruyter.com/document/doi/10.3139/104.111613/html.
- ↑ Kuhn, Thomas (1 October 2017). "Digitaler Zwilling" (in de). Informatik-Spektrum 40 (5): 440–444. doi:10.1007/s00287-017-1061-2. ISSN 0170-6012. http://link.springer.com/10.1007/s00287-017-1061-2.
- ↑ 76.0 76.1 76.2 Diaz-Flores, Ernesto; Meyer, Tim; Giorkallos, Alexis (2022), Beutel, Sascha; Lenk, Felix, eds., "Evolution of Artificial Intelligence-Powered Technologies in Biomedical Research and Healthcare" (in en), Smart Biolabs of the Future (Cham: Springer International Publishing) 182: 23–60, doi:10.1007/10_2021_189, ISBN 978-3-031-12581-2, https://link.springer.com/10.1007/10_2021_189. Retrieved 2024-04-23
- ↑ 77.0 77.1 Nikita, Saxena; Thakur, Garima; Jesubalan, Naveen G; Kulkarni, Amey; Yezhuvath, Vinesh B.; Rathore, Anurag S (1 August 2022). "AI-ML applications in bioprocessing: ML as an enabler of real time quality prediction in continuous manufacturing of mAbs" (in en). Computers & Chemical Engineering 164: 107896. doi:10.1016/j.compchemeng.2022.107896. https://linkinghub.elsevier.com/retrieve/pii/S0098135422002344.
- ↑ Jumper, John; Evans, Richard; Pritzel, Alexander; Green, Tim; Figurnov, Michael; Ronneberger, Olaf; Tunyasuvunakool, Kathryn; Bates, Russ et al. (26 August 2021). "Highly accurate protein structure prediction with AlphaFold" (in en). Nature 596 (7873): 583–589. doi:10.1038/s41586-021-03819-2. ISSN 0028-0836. PMC PMC8371605. PMID 34265844. https://www.nature.com/articles/s41586-021-03819-2.
- ↑ Biermann, Riekje; Niemeyer, Laura; Rösner, Laura; Ude, Christian; Lindner, Patrick; Bice, Ismet; Beutel, Sascha (1 March 2022). "Facilitated endospore detection for Bacillus spp. through automated algorithm‐based image processing" (in en). Engineering in Life Sciences 22 (3-4): 299–307. doi:10.1002/elsc.202100137. ISSN 1618-0240. PMC PMC8961035. PMID 35382541. https://onlinelibrary.wiley.com/doi/10.1002/elsc.202100137.
- ↑ Duong-Trung, Nghia; Born, Stefan; Kim, Jong Woo; Schermeyer, Marie-Therese; Paulick, Katharina; Borisyak, Maxim; Cruz-Bournazou, Mariano Nicolas; Werner, Thorben et al. (1 January 2023). "When bioprocess engineering meets machine learning: A survey from the perspective of automated bioprocess development" (in en). Biochemical Engineering Journal 190: 108764. doi:10.1016/j.bej.2022.108764. https://linkinghub.elsevier.com/retrieve/pii/S1369703X22004338.
- ↑ Hartmann, Fabian S.F.; Udugama, Isuru A.; Seibold, Gerd M.; Sugiyama, Hirokazu; Gernaey, Krist V. (1 November 2022). "Digital models in biotechnology: Towards multi-scale integration and implementation" (in en). Biotechnology Advances 60: 108015. doi:10.1016/j.biotechadv.2022.108015. https://linkinghub.elsevier.com/retrieve/pii/S0734975022001112.
- ↑ Oeing, Jonas; Henke, Fabian; Kockmann, Norbert (1 December 2021). "Machine Learning Based Suggestions of Separation Units for Process Synthesis in Process Simulation" (in en). Chemie Ingenieur Technik 93 (12): 1930–1936. doi:10.1002/cite.202100082. ISSN 0009-286X. https://onlinelibrary.wiley.com/doi/10.1002/cite.202100082.
- ↑ Zobel-Roos, Steffen; Stein, Dominik; Strube, Jochen (2 March 2018). "Evaluation of Continuous Membrane Chromatography Concepts with an Enhanced Process Simulation Approach" (in en). Antibodies 7 (1): 13. doi:10.3390/antib7010013. ISSN 2073-4468. PMC PMC6698847. PMID 31544865. https://www.mdpi.com/2073-4468/7/1/13.
- ↑ Ladd Effio, Christopher; Hahn, Tobias; Seiler, Julia; Oelmeier, Stefan A.; Asen, Iris; Silberer, Christine; Villain, Louis; Hubbuch, Jürgen (1 January 2016). "Modeling and simulation of anion-exchange membrane chromatography for purification of Sf9 insect cell-derived virus-like particles" (in en). Journal of Chromatography A 1429: 142–154. doi:10.1016/j.chroma.2015.12.006. https://linkinghub.elsevier.com/retrieve/pii/S0021967315017562.
- ↑ Boi, Cristiana; Dimartino, Simone; Sarti, Giulio C. (1 August 2007). "Modelling and simulation of affinity membrane adsorption" (in en). Journal of Chromatography A 1162 (1): 24–33. doi:10.1016/j.chroma.2007.02.008. https://linkinghub.elsevier.com/retrieve/pii/S0021967307002658.
- ↑ Torre, Luis de la; Neustock, Lars Thorben; Herring, George K.; Chacon, Jesus; Clemente, Felix J. Garcia; Hesselink, Lambertus (1 December 2020). "Automatic Generation and Easy Deployment of Digitized Laboratories". IEEE Transactions on Industrial Informatics 16 (12): 7328–7337. doi:10.1109/TII.2020.2977113. ISSN 1551-3203. https://ieeexplore.ieee.org/document/9018197/.
- ↑ Hatherly, P A; Jordan, S E; Cayless, A (1 July 2009). "Interactive screen experiments—innovative virtual laboratories for distance learners". European Journal of Physics 30 (4): 751–762. doi:10.1088/0143-0807/30/4/008. ISSN 0143-0807. https://iopscience.iop.org/article/10.1088/0143-0807/30/4/008.
- ↑ 88.0 88.1 88.2 Jones, Nicola (1 October 2018). "Simulated labs are booming" (in en). Nature 562 (7725): S5–S7. doi:10.1038/d41586-018-06831-1. ISSN 0028-0836. https://www.nature.com/articles/d41586-018-06831-1.
- ↑ Faulconer, Emily Kaye; Gruss, Amy B (1 May 2018). "A Review to Weigh the Pros and Cons of Online, Remote, and Distance Science Laboratory Experiences". The International Review of Research in Open and Distributed Learning 19 (2). doi:10.19173/irrodl.v19i2.3386. ISSN 1492-3831. http://www.irrodl.org/index.php/irrodl/article/view/3386.
- ↑ 90.0 90.1 Torre, Luis de la; Neustock, Lars Thorben; Herring, George K.; Chacon, Jesus; Clemente, Felix J. Garcia; Hesselink, Lambertus (1 December 2020). "Automatic Generation and Easy Deployment of Digitized Laboratories". IEEE Transactions on Industrial Informatics 16 (12): 7328–7337. doi:10.1109/TII.2020.2977113. ISSN 1551-3203. https://ieeexplore.ieee.org/document/9018197/.
- ↑ 91.0 91.1 91.2 91.3 Nakhle, Farid; Harfouche, Antoine L. (1 January 2023). "Extended reality gives digital agricultural biotechnology a new dimension" (in en). Trends in Biotechnology 41 (1): 1–5. doi:10.1016/j.tibtech.2022.09.005. https://linkinghub.elsevier.com/retrieve/pii/S0167779922002396.
- ↑ 92.0 92.1 Harfouche, Antoine L.; Nakhle, Farid (1 November 2020). "Creating Bioethics Distance Learning Through Virtual Reality" (in en). Trends in Biotechnology 38 (11): 1187–1192. doi:10.1016/j.tibtech.2020.05.005. PMC PMC7211661. PMID 32446631. https://linkinghub.elsevier.com/retrieve/pii/S0167779920301268.
- ↑ "Online Bachelor of Science in Biological Sciences". ASU Online. Arizona State University. https://asuonline.asu.edu/online-degree-programs/undergraduate/biological-sciences-bs/.
- ↑ Stefani, Caroline; Lacy-Hulbert, Adam; Skillman, Thomas (1 October 2018). "ConfocalVR: Immersive Visualization for Confocal Microscopy" (in en). Journal of Molecular Biology 430 (21): 4028–4035. doi:10.1016/j.jmb.2018.06.035. PMC PMC6805139. PMID 29949752. https://linkinghub.elsevier.com/retrieve/pii/S0022283618306648.
- ↑ Nee, A.Y.C.; Ong, S.K.; Chryssolouris, G.; Mourtzis, D. (2012). "Augmented reality applications in design and manufacturing" (in en). CIRP Annals 61 (2): 657–679. doi:10.1016/j.cirp.2012.05.010. https://linkinghub.elsevier.com/retrieve/pii/S0007850612002090.
- ↑ Mourtzis, Dimitris; Siatras, Vasileios; Angelopoulos, John (8 March 2020). "Real-Time Remote Maintenance Support Based on Augmented Reality (AR)" (in en). Applied Sciences 10 (5): 1855. doi:10.3390/app10051855. ISSN 2076-3417. https://www.mdpi.com/2076-3417/10/5/1855.
- ↑ Lamberti, Fabrizio; Manuri, Federico; Sanna, Andrea; Paravati, Gianluca; Pezzolla, Pietro; Montuschi, Paolo (1 December 2014). "Challenges, Opportunities, and Future Trends of Emerging Techniques for Augmented Reality-Based Maintenance". IEEE Transactions on Emerging Topics in Computing 2 (4): 411–421. doi:10.1109/TETC.2014.2368833. ISSN 2168-6750. http://ieeexplore.ieee.org/document/7004838/.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. A fair amount of grammar and punctuation was updated from the original. In some cases important information was missing from the references, and that information was added.